- Record: found
- Abstract: found
- Article: found
Anti-nucleocapsid antibody levels following initial and repeat SARS-CoV-2 infections in a cohort of long-term care facility residents in England (VIVALDI)
Read this article at
Abstract
Background
We have previously demonstrated that older residents of long-term care facilities (LTCF) in the UK show levels of anti-spike antibodies that are comparable to the general population following primary series and booster vaccination for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, data on the humoral response to other SARS-CoV-2 proteins associated with natural infection are scarce in this vulnerable population.
Methods
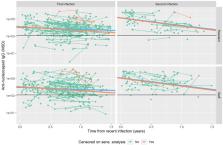
We measured quantitative levels of anti-nucleocapsid antibodies in blood samples taken from LTCF residents and staff after initial and repeat SARS-CoV-2 infections, between December 2020 and March 2023. Data on SARS-CoV-2 infection and vaccination were obtained through linkage to national datasets. Linear mixed effects models were used to investigate anti-nucleocapsid antibody levels, using log10 scale, in relation to time from most recent infection. This included evaluation of associations between repeat infection, staff/resident status, age, sex, Omicron infection and vaccination history and peak antibody level and slope of decline with time.
Results
We analysed 405 antibody observations from 220 residents and 396 observations from 215 staff. Repeat infection was associated with 8.5-fold (95%CI 4.9-14.8-fold) higher initial (peak) median anti-nucleocapsid antibody level, with steeper subsequent slope of decline. There were no significant differences in antibody level associated with resident (vs. staff) status or age, but Omicron infection was associated with 3.6-fold (95%CI 2.4–5.4-fold) higher levels. There was stronger evidence of waning of antibody levels over time in a sensitivity analysis in which observations were censored in cases with suspected undetected repeat infection.
Plain Language Summary
COVID-19 had a severe impact on care homes in the UK early in the pandemic. However, deaths and disease caused by the SARS-CoV-2 virus have decreased over time following successful introduction of vaccinations and resistance linked to prior infection. There has been a lot of research carried out on the body's immune response to the viral spike protein, which was used to create vaccines against the virus. Less is known about our immune response to other proteins produced by the virus, such as nucleocapsid, which have not been used in current vaccines.
We evaluated antibody levels against the viral nucleocapsid protein in older care home residents following initial and repeat SARS-CoV-2 infection and compared these values to those observed in younger care home staff. This was done through a large established cohort study, in which residents and staff of participating homes could volunteer to provide blood samples for analysis.
We found similar levels of antibody levels among staff and older residents of care homes. These findings are in line with previous studies, in which we have shown that care home residents who survive SARS-CoV-2 infection can develop robust immunity. Higher peak antibody levels were observed following repeat infection in both residents and staff.
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: found
The duration, dynamics and determinants of SARS-CoV-2 antibody responses in individual healthcare workers
- Record: found
- Abstract: found
- Article: found
Prevalence and duration of detectable SARS-CoV-2 nucleocapsid antibodies in staff and residents of long-term care facilities over the first year of the pandemic (VIVALDI study): prospective cohort study in England

- Record: found
- Abstract: found
- Article: found