- Record: found
- Abstract: found
- Article: found
A review on machine learning approaches and trends in drug discovery

Read this article at
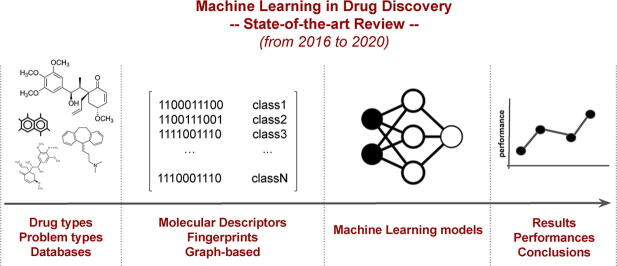
Graphical abstract
Highlights
Abstract
Drug discovery aims at finding new compounds with specific chemical properties for the treatment of diseases. In the last years, the approach used in this search presents an important component in computer science with the skyrocketing of machine learning techniques due to its democratization. With the objectives set by the Precision Medicine initiative and the new challenges generated, it is necessary to establish robust, standard and reproducible computational methodologies to achieve the objectives set. Currently, predictive models based on Machine Learning have gained great importance in the step prior to preclinical studies. This stage manages to drastically reduce costs and research times in the discovery of new drugs. This review article focuses on how these new methodologies are being used in recent years of research. Analyzing the state of the art in this field will give us an idea of where cheminformatics will be developed in the short term, the limitations it presents and the positive results it has achieved. This review will focus mainly on the methods used to model the molecular data, as well as the biological problems addressed and the Machine Learning algorithms used for drug discovery in recent years.
Related collections
Most cited references124
- Record: found
- Abstract: not found
- Article: not found
A tutorial on support vector regression
- Record: found
- Abstract: found
- Article: found
The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups
- Record: found
- Abstract: found
- Article: not found