- Record: found
- Abstract: found
- Article: found
The role of microbiome-host interactions in the development of Alzheimer´s disease
Read this article at
Abstract
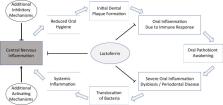
Alzheimer`s disease (AD) is the most prevalent cause of dementia. It is often assumed that AD is caused by an aggregation of extracellular beta-amyloid and intracellular tau-protein, supported by a recent study showing reduced brain amyloid levels and reduced cognitive decline under treatment with a beta-amyloid-binding antibody. Confirmation of the importance of amyloid as a therapeutic target notwithstanding, the underlying causes of beta-amyloid aggregation in the human brain, however, remain to be elucidated. Multiple lines of evidence point towards an important role of infectious agents and/or inflammatory conditions in the etiology of AD. Various microorganisms have been detected in the cerebrospinal fluid and brains of AD-patients and have thus been hypothesized to be linked to the development of AD, including Porphyromonas gingivalis (PG) and Spirochaetes. Intriguingly, these microorganisms are also found in the oral cavity under normal physiological conditions, which is often affected by multiple pathologies like caries or tooth loss in AD patients. Oral cavity pathologies are mostly accompanied by a compositional shift in the community of oral microbiota, mainly affecting commensal microorganisms and referred to as ‘dysbiosis’. Oral dysbiosis seems to be at least partly mediated by key pathogens such as PG, and it is associated with a pro-inflammatory state that promotes the destruction of connective tissue in the mouth, possibly enabling the translocation of pathogenic microbiota from the oral cavity to the nervous system. It has therefore been hypothesized that dysbiosis of the oral microbiome may contribute to the development of AD. In this review, we discuss the infectious hypothesis of AD in the light of the oral microbiome and microbiome-host interactions, which may contribute to or even cause the development of AD. We discuss technical challenges relating to the detection of microorganisms in relevant body fluids and approaches for avoiding false-positives, and introduce the antibacterial protein lactoferrin as a potential link between the dysbiotic microbiome and the host inflammatory reaction.
Related collections
Most cited references158

- Record: found
- Abstract: found
- Article: found
Metagenomic biomarker discovery and explanation
- Record: found
- Abstract: found
- Article: not found
Lecanemab in Early Alzheimer’s Disease
- Record: found
- Abstract: found
- Article: not found