- Record: found
- Abstract: found
- Article: found
The effect of ecosystem biodiversity on virus genetic diversity depends on virus species: A study of chiltepin-infecting begomoviruses in Mexico
Read this article at
Abstract
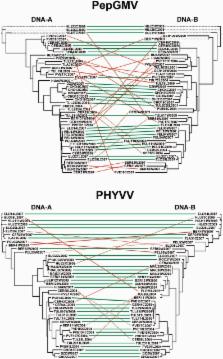
Current declines in biodiversity put at risk ecosystem services that are fundamental for human welfare. Increasing evidence indicates that one such service is the ability to reduce virus emergence. It has been proposed that the reduction of virus emergence occurs at two levels: through a reduction of virus prevalence/transmission and, as a result of these epidemiological changes, through a limitation of virus genetic diversity. Although the former mechanism has been studied in a few host-virus interactions, very little is known about the association between ecosystem biodiversity and virus genetic diversity. To address this subject, we estimated genetic diversity, synonymous and non-synonymous nucleotide substitution rates, selection pressures, and frequency of recombinants and re-assortants in populations of Pepper golden mosaic virus (PepGMV) and Pepper huasteco yellow vein virus (PHYVV) that infect chiltepin plants in Mexico. We then analyzed how these parameters varied according to the level of habitat anthropization, which is the major cause of biodiversity loss. Our results indicated that genetic diversity of PepGMV (but not of PHYVV) populations increased with the loss of biodiversity at higher levels of habitat anthropization. This was mostly the consequence of higher rates of synonymous nucleotide substitutions, rather than of adaptive selection. The frequency of recombinants and re-assortants was higher in PepGMV populations infecting wild chiltepin than in those infecting cultivated ones, suggesting that genetic exchange is not the main mechanism for generating genetic diversity in PepGMV populations. These findings provide evidence that biodiversity may modulate the genetic diversity of plant viruses, but it may differentially affect even two closely related viruses. Our analyses may contribute to understanding the factors involved in virus emergence.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
DnaSP, DNA polymorphism analyses by the coalescent and other methods.

- Record: found
- Abstract: found
- Article: found
Jane: a new tool for the cophylogeny reconstruction problem
- Record: found
- Abstract: found
- Article: not found