- Record: found
- Abstract: found
- Article: not found
COVID-19 SignSym: a fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model
Read this article at
Abstract
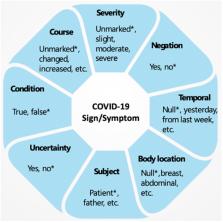
The COVID-19 pandemic swept across the world rapidly, infecting millions of people. An efficient tool that can accurately recognize important clinical concepts of COVID-19 from free text in electronic health records (EHRs) will be valuable to accelerate COVID-19 clinical research. To this end, this study aims at adapting the existing CLAMP natural language processing tool to quickly build COVID-19 SignSym, which can extract COVID-19 signs/symptoms and their 8 attributes (body location, severity, temporal expression, subject, condition, uncertainty, negation, and course) from clinical text. The extracted information is also mapped to standard concepts in the Observational Medical Outcomes Partnership common data model. A hybrid approach of combining deep learning-based models, curated lexicons, and pattern-based rules was applied to quickly build the COVID-19 SignSym from CLAMP, with optimized performance. Our extensive evaluation using 3 external sites with clinical notes of COVID-19 patients, as well as the online medical dialogues of COVID-19, shows COVID-19 SignSym can achieve high performance across data sources. The workflow used for this study can be generalized to other use cases, where existing clinical natural language processing tools need to be customized for specific information needs within a short time. COVID-19 SignSym is freely accessible to the research community as a downloadable package ( https://clamp.uth.edu/covid/nlp.php) and has been used by 16 healthcare organizations to support clinical research of COVID-19.
Related collections
Most cited references7
- Record: found
- Abstract: found
- Article: not found
Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications.

- Record: found
- Abstract: found
- Article: found
CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines
- Record: found
- Abstract: found
- Article: not found
