- Record: found
- Abstract: found
- Article: found
Neutrophil proteases are protective against SARS-CoV-2 by degrading the spike protein and dampening virus-mediated inflammation

Read this article at
Abstract
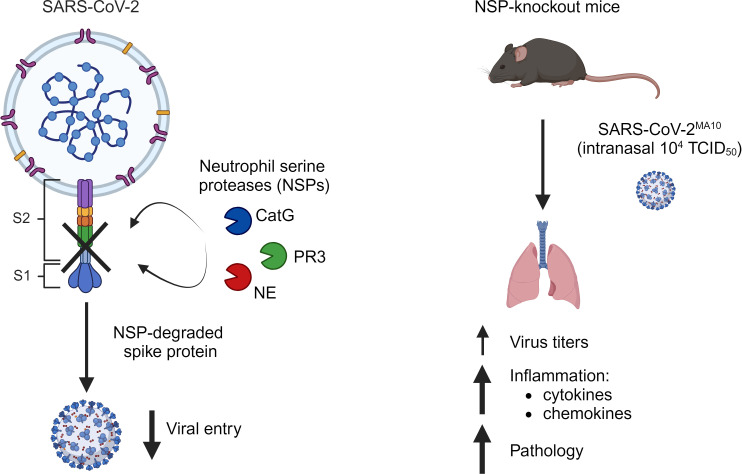
Studies on severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) have highlighted the crucial role of host proteases for viral replication and the immune response. The serine proteases furin and TMPRSS2 and lysosomal cysteine proteases facilitate viral entry by limited proteolytic processing of the spike (S) protein. While neutrophils are recruited to the lungs during COVID-19 pneumonia, little is known about the role of the neutrophil serine proteases (NSPs) cathepsin G (CatG), elastase (NE), and proteinase 3 (PR3) on SARS-CoV-2 entry and replication. Furthermore, the current paradigm is that NSPs may contribute to the pathogenesis of severe COVID-19. Here, we show that these proteases cleaved the S protein at multiple sites and abrogated viral entry and replication in vitro. In mouse models, CatG significantly inhibited viral replication in the lung. Importantly, lung inflammation and pathology were increased in mice deficient in NE and/or CatG. These results reveal that NSPs contribute to innate defenses against SARS-CoV-2 infection via proteolytic inactivation of the S protein and that NE and CatG limit lung inflammation in vivo. We conclude that therapeutic interventions aiming to reduce the activity of NSPs may interfere with viral clearance and inflammation in COVID-19 patients.
Abstract
Abstract
Neutrophil granule serine proteases cleave the spike protein and are mainly protective in early SARS-CoV-2 infection in mice.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
- Record: found
- Abstract: found
- Article: not found
A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells
- Record: found
- Abstract: found
- Article: not found