- Record: found
- Abstract: found
- Article: not found
Discovery of novel non-competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)

Abstract
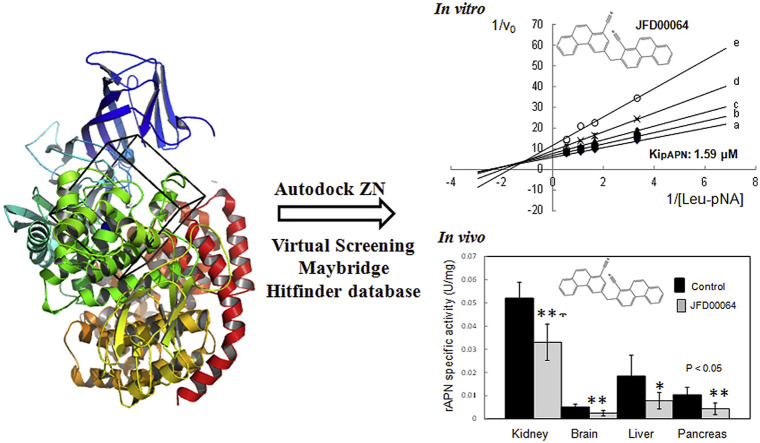
Neutral metallo-aminopeptidase (APN) catalyzes the cleavage of neutral and basic amino acids from the N-terminus of protein or peptide substrates. APN expression is dysregulated in inflammatory diseases as well as in several types of cancer. Therefore, inhibitors of APN may be effective against cancer and inflammation. By virtual screening and enzymatic assays, we identified three non-competitive inhibitors (α > 1) of the porcine and human APN with K i values in the μM range. These non-peptidic compounds lack the classical zinc-binding groups (ZBG) present in most of the APN inhibitors. Molecular docking simulations suggested the novel inhibitors suppress APN activity by an alternative mechanism to Zn coordination: they interacted with residues comprising the S1 and S5′ subsites of APN. Of note, these compounds also inhibited the porcine aminopeptidase A (pAPA) using a competitive inhibition mode. This indicated differences in the binding mode of these compounds with APN and APA. Based on sequence and structural analyses, we predicted the significance of targeting human APN residues: Ala-351, Arg-442, Ala-474, Phe-896 and Asn-900 for improving the selectivity of the identified compounds. Remarkably, the intraperitoneal injection of compounds BTB07018 and JFD00064 inhibited APN activity in rat brain, liver and kidney indicating good bio-distribution of these inhibitors in vivo. These data reinforce the idea of designing novel APN inhibitors based on lead compounds without ZBG.
Graphical abstract
Highlights
-
•
We identified three non-competitive inhibitors of the human and porcine APN.
-
•
These compounds lack the classical zinc-binding groups of the APN inhibitors.
-
•
We proposed these molecules block APN by an alternative mechanism to Zn chelation.
-
•
All the inhibitors interact with APN residues comprising the S1 and S5′ subsites.
-
•
Two compounds blocked the APN activity in the brain, liver and kidney of rats.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions.

- Record: found
- Abstract: found
- Article: found
Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors
- Record: found
- Abstract: found
- Article: not found