- Record: found
- Abstract: found
- Article: found
Lentiviral Vectors for Delivery of Gene-Editing Systems Based on CRISPR/Cas: Current State and Perspectives
Read this article at
Abstract
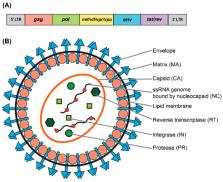
CRISPR/Cas technology has revolutionized the fields of the genome- and epigenome-editing by supplying unparalleled control over genomic sequences and expression. Lentiviral vector (LV) systems are one of the main delivery vehicles for the CRISPR/Cas systems due to ( i) its ability to carry bulky and complex transgenes and ( ii) sustain robust and long-term expression in a broad range of dividing and non-dividing cells in vitro and in vivo. It is thus reasonable that substantial effort has been allocated towards the development of the improved and optimized LV systems for effective and accurate gene-to-cell transfer of CRISPR/Cas tools. The main effort on that end has been put towards the improvement and optimization of the vector’s expression, development of integrase-deficient lentiviral vector (IDLV), aiming to minimize the risk of oncogenicity, toxicity, and pathogenicity, and enhancing manufacturing protocols for clinical applications required large-scale production. In this review, we will devote attention to ( i) the basic biology of lentiviruses, and ( ii) recent advances in the development of safer and more efficient CRISPR/Cas vector systems towards their use in preclinical and clinical applications. In addition, we will discuss in detail the recent progress in the repurposing of CRISPR/Cas systems related to base-editing and prime-editing applications.
Related collections
Most cited references81
- Record: found
- Abstract: found
- Article: not found
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
- Record: found
- Abstract: not found
- Article: not found
Improved vectors and genome-wide libraries for CRISPR screening.
- Record: found
- Abstract: found
- Article: not found