- Record: found
- Abstract: found
- Article: found
Circular RNAs in cell cycle regulation: Mechanisms to clinical significance
Read this article at
Abstract
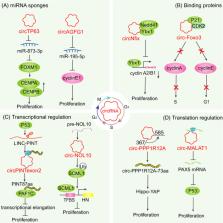
Circular RNAs (circRNAs), a type of non‐coding RNA, are single‐stranded circularized molecules characterized by high abundance, evolutionary conservation and cell development‐ and tissue‐specific expression. A large body of studies has found that circRNAs exert a wide variety of functions in diverse biological processes, including cell cycle. The cell cycle is controlled by the coordinated activation and deactivation of cell cycle regulators. CircRNAs exert mutifunctional roles by regulating gene expression via various mechanisms. However, the functional relevance of circRNAs and cell cycle regulation largely remains to be elucidated. Herein, we briefly describe the biogenesis and mechanistic models of circRNAs and summarize their functions and mechanisms in the regulation of critical cell cycle modulators, including cyclins, cyclin‐dependent kinases and cyclin‐dependent kinase inhibitors. Moreover, we highlight the participation of circRNAs in cell cycle‐related signalling pathways and the clinical value of circRNAs as promising biomarkers or therapeutic targets in diseases related to cell cycle disorder.
Abstract
CircRNAs exert a wide variety of functions in diverse biological processes, including cell cycle. The cell cycle is controlled by the coordinated activation and deactivation of cell cycle regulators, including cyclins, cyclin‐dependent kinases and cyclin‐dependent kinase inhibitors. This review summarizes the functions and mechanisms of circRNAs in cell cycle regulation and highlights the clinical value of circRNAs as promising biomarkers or therapeutic targets.
Related collections
Most cited references126
- Record: found
- Abstract: found
- Article: not found
The biogenesis, biology and characterization of circular RNAs
- Record: found
- Abstract: found
- Article: not found
Circular RNAs are abundant, conserved, and associated with ALU repeats.
- Record: found
- Abstract: found
- Article: not found
