- Record: found
- Abstract: found
- Article: found
CircEHD2, CircNETO2 and CircEGLN3 as Diagnostic and Prognostic Biomarkers for Patients with Renal Cell Carcinoma
Read this article at
Abstract
Simple Summary
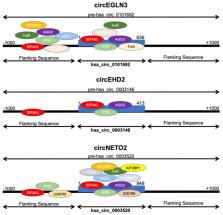
Circular RNA (circRNA) plays an important role in cancer, but little is known about its role in clear cell renal cell carcinoma (ccRCC). The study was designed to analyze the role of circRNAs in ccRCC. We show that circEHD2, circENGLN3, and circNETO2 are upregulated in ccRCC compared with non-malignant renal tissue. Increased circEHD2 levels were significant and independent predictors of progression-free and cancer-specific survival of ccRCC patients. Thus, the analysis of circRNAs may be of diagnostic and prognostic relevance in patients with ccRCC.
Abstract
Background: Circular RNA (circRNA) plays an important role in the carcinogenesis of various tumors. It is assumed that circRNAs have a high tissue and tumor specificity, thus they are discussed as cancer biomarkers. The knowledge about circRNAs in clear cell renal carcinoma (ccRCC) is limited so far, and thus we studied the expression profile of seven circRNAs (circCOL5A1, circEHD2, circEDEM2, circEGNL3, circNETO2, circSCARB1, circSOD2) in a cohort of ccRCC patients. Methods: Fresh-frozen normal and cancerous tissues were prospectively collected from patients with ccRCC undergoing partial/radical nephrectomy. Total RNA was isolated from 121 ccRCC and 91 normal renal tissues, and the circRNA expression profile was determined using quantitative real-time PCR. Results: circEHD2, circENGLN3, and circNETO2 were upregulated in ccRCC compared with non-malignant renal tissue. circENGLN3 expression was highly discriminative between normal and cancerous tissue. None of the circRNAs was correlated with clinicopathological parameters. High circEHD2 and low circNETO2 levels were an independent predictor of a shortened progression-free survival, cancer-specific survival, and overall survival in patients with ccRCC undergoing nephrectomy. Conclusions: The analysis of circRNAs may provide diagnostic and prognostic information. Thus, circRNAs could help to optimize the individual treatment and ultimately improve ccRCC patients’ survival.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs.
- Record: found
- Abstract: found
- Article: not found
Sunitinib Alone or after Nephrectomy in Metastatic Renal-Cell Carcinoma

- Record: found
- Abstract: found
- Article: found