- Record: found
- Abstract: found
- Article: found
Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis
Read this article at
Abstract
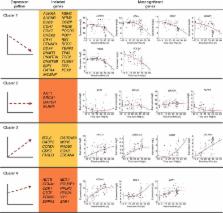
Aneuploidies are prevalent in the human embryo and impair proper development, leading to cell cycle arrest. Recent advances in imaging and molecular and genetic analyses are postulated as promising strategies to unveil the mechanisms involved in aneuploidy generation. Here we combine time-lapse, complete chromosomal assessment and single-cell RT–qPCR to simultaneously obtain information from all cells that compose a human embryo until the approximately eight-cell stage ( n=85). Our data indicate that the chromosomal status of aneuploid embryos ( n=26), including those that are mosaic ( n=3), correlates with significant differences in the duration of the first mitotic phase when compared with euploid embryos ( n=28). Moreover, gene expression profiling suggests that a subset of genes is differentially expressed in aneuploid embryos during the first 30 h of development. Thus, we propose that the chromosomal fate of an embryo is likely determined as early as the pronuclear stage and may be predicted by a 12-gene transcriptomic signature.
Abstract
 Aneuploidy may be fatal for the embryo, hence predicting its occurrence is important
for successful
in vitro fertilization. Here the authors monitor development of human preimplantation embryos
in real-time and correlate the blastomere ploidy with cleavage dynamics and gene expression,
identifying 12-transcript signature that determines ploidy.
Aneuploidy may be fatal for the embryo, hence predicting its occurrence is important
for successful
in vitro fertilization. Here the authors monitor development of human preimplantation embryos
in real-time and correlate the blastomere ploidy with cleavage dynamics and gene expression,
identifying 12-transcript signature that determines ploidy.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
NIH Image to ImageJ: 25 years of image analysis.
- Record: found
- Abstract: found
- Article: not found
Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing.
- Record: found
- Abstract: found
- Article: not found