- Record: found
- Abstract: found
- Article: found
Complete chloroplast genomes of the hemiparasitic genus Cymbaria: Insights into comparative analysis, development of molecular markers, and phylogenetic relationships

Read this article at
Abstract
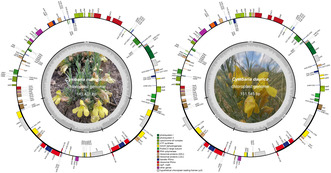
The hemiparasitic tribe Cymbarieae (Orobanchaceae) plays a crucial role in elucidating the initial stage of the transition from autotrophism to heterotrophism. However, the complete chloroplast genome of the type genus Cymbaria has yet to be reported. In addition, the traditional Mongolian medicine Cymbaria daurica is frequently subjected to adulteration or substitution because of the minor morphological differences with Cymbaria mongolica. In this study, the complete chloroplast genomes of the two Cymbaria species were assembled and annotated, and those of other published 52 Orobanchaceae species were retrieved for comparative analyses. We found that the Cymbaria chloroplast genomes are characterized by pseudogenization or loss of stress‐relevant genes ( ndh) and a unique rbcL– matK inversion. Unlike the high variability observed in holoparasites, Cymbaria and other hemiparasites exhibit high similarity to autotrophs in genome size, guanine–cytosine (GC) content, and intact genes. Notably, four pairs of specific DNA barcodes were developed and validated to distinguish the medicinal herb from its adulterants. Phylogenetic analyses revealed that the genus Cymbaria and the Schwalbea– Siphonostegia clade are grouped into the tribe Cymbarieae, which forms a sister clade to the remaining Orobanchaceae parasitic lineages. Moreover, the diversification of monophyletic Cymbaria occurred during the late Miocene (6.72 Mya) in the Mongol–Chinese steppe region. Our findings provide valuable genetic resources for studying the phylogeny of Orobanchaceae and plant parasitism, and genetic tools to validate the authenticity of the traditional Mongolian medicine “ Xinba.”
Abstract
Related collections
Most cited references95

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: not found