- Record: found
- Abstract: found
- Article: found
Fifteen-Year Application of Manure and Chemical Fertilizers Differently Impacts Soil ARGs and Microbial Community Structure
Read this article at
Abstract
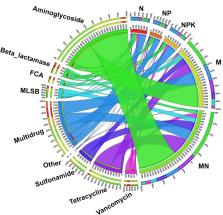
Manure, which contains large amounts of antibiotics and antibiotic resistance genes (ARGs), is widely used in agricultural soils and may lead to the evolution and dispersal of ARGs in the soil environment. In the present study, soils that received manure or chemical fertilizers for 15 years were sampled on the North China Plain (NCP), which is one of the primary areas of intensive agriculture in China. High-throughput quantitative PCR and sequencing technologies were employed to assess the effects of long-term manure or chemical fertilizer application on the distribution of ARGs and microbial communities. A total of 114 unique ARGs were successfully amplified from all soil samples. Manure application markedly increased the relative abundance and detectable numbers of ARGs, with up to 0.23 copies/16S rRNA gene and 81 unique ARGs. The increased abundance of ARGs in manure-fertilized soil was mainly due to the manure increasing the abundance of indigenous soil ARGs. In contrast, chemical fertilizers only moderately affected the diversity of ARGs and had no significant effect on the relative abundance of the total ARGs. In addition, manure application increased the abundance of mobile genetic elements (MGEs), which were significantly and positively correlated with most types of ARGs, indicating that horizontal gene transfer via MGEs may play an important role in the spread of ARGs. Furthermore, the application of manure and chemical fertilizers significantly affected microbial community structure, and variation partitioning analysis showed that microbial community shifts represented the major driver shaping the antibiotic resistome. Taken together, our results provide insight into the long-term effects of manure and chemical fertilization on the dissemination of ARGs in intensive agricultural ecosystems.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Tackling antibiotic resistance: the environmental framework.
- Record: found
- Abstract: found
- Article: not found
Rhizosphere microbiome assemblage is affected by plant development.
- Record: found
- Abstract: found
- Article: not found