- Record: found
- Abstract: found
- Article: found
Development of an efficient gene-targeting system for elucidating infection mechanisms of the fungal pathogen Trichosporon asahii
Read this article at
Abstract
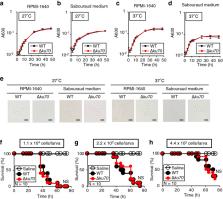
Trichosporon asahii is a pathogenic fungus that causes severe, deep-seated fungal infections in neutropenic patients . Elucidating the infection mechanisms of T. asahii based on genetic studies requires a specific gene-targeting system. Here, we established an efficient gene-targeting system in a highly pathogenic T. asahii strain identified using the silkworm infection model. By comparing the pathogenicity of T. asahii clinical isolates in a silkworm infection model, T. asahii MPU129 was identified as a highly pathogenic strain. Using an Agrobacterium tumefaciens-mediated gene transfer system, we obtained a T. asahii MPU129 mutant lacking the ku70 gene, which encodes the Ku70 protein involved in the non-homologous end-joining repair of DNA double-strand breaks. The ku70 gene-deficient mutant showed higher gene-targeting efficiency than the wild-type strain for constructing a mutant lacking the cnb1 gene, which encodes the beta-subunit of calcineurin. The cnb1 gene-deficient mutant showed reduced pathogenicity against silkworms compared with the parental strain. These results suggest that an efficient gene-targeting system in a highly pathogenic T. asahii strain is a useful tool for elucidating the molecular mechanisms of T. asahii infection.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Current knowledge of Trichosporon spp. and Trichosporonosis.
- Record: found
- Abstract: found
- Article: not found
The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus.
- Record: found
- Abstract: found
- Article: not found
Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.