- Record: found
- Abstract: found
- Article: found
The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA
Read this article at
Abstract
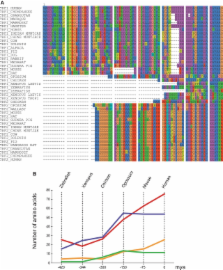
TRF1 and TRF2 are key proteins in human telomeres, which, despite their similarities, have different behaviors upon DNA binding. Previous work has shown that unlike TRF1, TRF2 condenses telomeric, thus creating consequential negative torsion on the adjacent DNA, a property that is thought to lead to the stimulation of single-strand invasion and was proposed to favor telomeric DNA looping. In this report, we show that these activities, originating from the central TRFH domain of TRF2, are also displayed by the TRFH domain of TRF1 but are repressed in the full-length protein by the presence of an acidic domain at the N-terminus. Strikingly, a similar repression is observed on TRF2 through the binding of a TERRA-like RNA molecule to the N-terminus of TRF2. Phylogenetic and biochemical studies suggest that the N-terminal domains of TRF proteins originate from a gradual extension of the coding sequences of a duplicated ancestral gene with a consequential progressive alteration of the biochemical properties of these proteins. Overall, these data suggest that the N-termini of TRF1 and TRF2 have evolved to finely regulate their ability to condense DNA.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny.
- Record: found
- Abstract: found
- Article: not found
TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres.
- Record: found
- Abstract: found
- Article: not found