- Record: found
- Abstract: found
- Article: found
Therapeutic mitigation of measles-like immune amnesia and exacerbated disease after prior respiratory virus infections in ferrets
Read this article at
Abstract
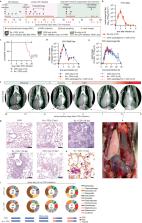
Measles cases have surged pre-COVID-19 and the pandemic has aggravated the problem. Most measles-associated morbidity and mortality arises from destruction of pre-existing immune memory by measles virus (MeV), a paramyxovirus of the morbillivirus genus. Therapeutic measles vaccination lacks efficacy, but little is known about preserving immune memory through antivirals and the effect of respiratory disease history on measles severity. We use a canine distemper virus (CDV)-ferret model as surrogate for measles and employ an orally efficacious paramyxovirus polymerase inhibitor to address these questions. A receptor tropism-intact recombinant CDV with low lethality reveals an 8-day advantage of antiviral treatment versus therapeutic vaccination in maintaining immune memory. Infection of female ferrets with influenza A virus (IAV) A/CA/07/2009 (H1N1) or respiratory syncytial virus (RSV) four weeks pre-CDV causes fatal hemorrhagic pneumonia with lung onslaught by commensal bacteria. RNAseq identifies CDV-induced overexpression of trefoil factor (TFF) peptides in the respiratory tract, which is absent in animals pre-infected with IAV. Severe outcomes of consecutive IAV/CDV infections are mitigated by oral antivirals even when initiated late. These findings validate the morbillivirus immune amnesia hypothesis, define measles treatment paradigms, and identify priming of the TFF axis through prior respiratory infections as risk factor for exacerbated morbillivirus disease.
Abstract
Measles virus infection causes immunosuppression and it’s unclear whether this can be prevented by antivirals. Here, using a canine distemper virus ferret model, the authors show that measles-like immune amnesia and lethal exacerbated respiratory disease after consecutive respiratory virus infections can be mitigated by oral antiviral therapy initiated at the onset of primary clinical signs.
Related collections
Most cited references50

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found