- Record: found
- Abstract: found
- Article: found
Tuned SMC Arms Drive Chromosomal Loading of Prokaryotic Condensin
Read this article at
Summary
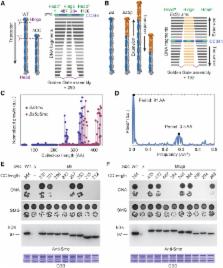
SMC proteins support vital cellular processes in all domains of life by organizing chromosomal DNA. They are composed of ATPase “head” and “hinge“ dimerization domains and a connecting coiled-coil “arm.” Binding to a kleisin subunit creates a closed tripartite ring, whose ∼47-nm-long SMC arms act as barrier for DNA entrapment. Here, we uncover another, more active function of the bacterial Smc arm. Using high-throughput genetic engineering, we resized the arm in the range of 6–60 nm and found that it was functional only in specific length regimes following a periodic pattern. Natural SMC sequences reflect these length constraints. Mutants with improper arm length or peptide insertions in the arm efficiently target chromosomal loading sites and hydrolyze ATP but fail to use ATP hydrolysis for relocation onto flanking DNA. We propose that SMC arms implement force transmission upon nucleotide hydrolysis to mediate DNA capture or loop extrusion.
Graphical Abstract
Highlights
-
•
Short and long but not intermediate-length Smc coiled-coil arms are functional
-
•
Smc complexes with improper arms accumulate at chromosomal loading sites
-
•
Smc arms are functional units linking ATP hydrolysis to an essential DNA transaction
-
•
Pro- and eukaryotic SMC sequences reflect similar periodic length constraints
Abstract
By engineering a series of Smc proteins with shorter or longer coiled-coil arms, Bürmann et al. elucidate a critical role for the arms’ super-helical nature. Improper arms support chromosomal targeting but fail to link ATP hydrolysis to chromosomal loading. Smc arms are proposed to implement force transmission upon nucleotide hydrolysis.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
Molecular architecture of SMC proteins and the yeast cohesin complex.

- Record: found
- Abstract: found
- Article: found
Self-organization of domain structures by DNA-loop-extruding enzymes
- Record: found
- Abstract: found
- Article: not found
