- Record: found
- Abstract: found
- Article: found
Enterohemorrhagic Escherichia coli pathogenesis: role of Long polar fimbriae in Peyer’s patches interactions
Read this article at
Abstract
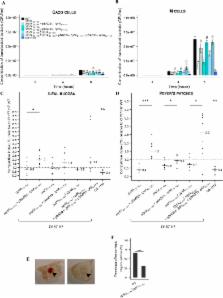
Enterohemorrhagic Escherichia coli (EHEC) are major food-borne pathogens whose survival and virulence in the human digestive tract remain unclear owing to paucity of relevant models. EHEC interact with the follicle-associated epithelium of Peyer’s patches of the distal ileum and translocate across the intestinal epithelium via M-cells, but the underlying molecular mechanisms are still unknown. Here, we investigated the involvement of Long polar fimbriae (Lpf) in EHEC pathogenesis. Of the 236 strains tested, a significant association was observed between the presence of lpf operons and pathogenicity. In sophisticated in vitro models of the human gastro-intestinal tract, lpf expression was induced during transit through the simulated stomach and small intestine, but not in the colonic compartment. To investigate the involvement of Lpf in EHEC pathogenesis, lpf isogenic mutants and their relative trans-complemented strains were generated. Translocation across M-cells, interactions with murine ileal biopsies containing Peyer’s patches and the number of hemorrhagic lesions were significantly reduced with the lpf mutants compared to the wild-type strain. Complementation of lpf mutants fully restored the wild-type phenotypes. Our results indicate that (i) EHEC might colonize the terminal ileum at the early stages of infection, (ii) Lpf are an important player in the interactions with Peyer’s patches and M-cells, and could contribute to intestinal colonization.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
The role of pH in determining the species composition of the human colonic microbiota.
- Record: found
- Abstract: found
- Article: not found
Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response.
- Record: found
- Abstract: found
- Article: not found