- Record: found
- Abstract: found
- Article: found
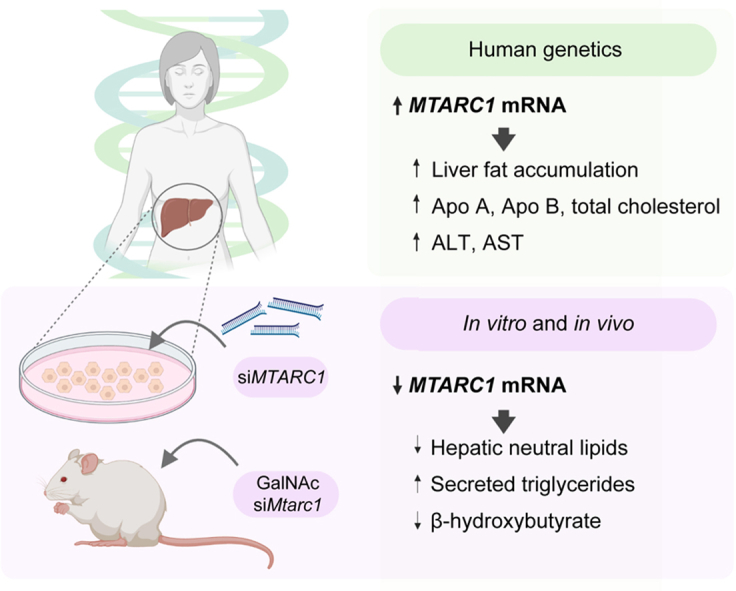
Hepatocyte mARC1 promotes fatty liver disease
Read this article at
Abstract
Background & Aims
Non-alcoholic fatty liver disease (NAFLD) has a prevalence of ∼25% worldwide, with significant public health consequences yet few effective treatments. Human genetics can help elucidate novel biology and identify targets for new therapeutics. Genetic variants in mitochondrial amidoxime-reducing component 1 ( MTARC1) have been associated with NAFLD and liver-related mortality; however, its pathophysiological role and the cell type(s) mediating these effects remain unclear. We aimed to investigate how MTARC1 exerts its effects on NAFLD by integrating human genetics with in vitro and in vivo studies of mARC1 knockdown.
Methods
Analyses including multi-trait colocalisation and Mendelian randomisation were used to assess the genetic associations of MTARC1. In addition, we established an in vitro long-term primary human hepatocyte model with metabolic readouts and used the Gubra Amylin NASH (GAN)-diet non-alcoholic steatohepatitis mouse model treated with hepatocyte-specific N-acetylgalactosamine (GalNAc)–siRNA to understand the in vivo impacts of MTARC1.
Results
We showed that genetic variants within the MTARC1 locus are associated with liver enzymes, liver fat, plasma lipids, and body composition, and these associations are attributable to the same causal variant ( p.A165T, rs2642438 G>A), suggesting a shared mechanism. We demonstrated that increased MTARC1 mRNA had an adverse effect on these traits using Mendelian randomisation, implying therapeutic inhibition of mARC1 could be beneficial. In vitro mARC1 knockdown decreased lipid accumulation and increased triglyceride secretion, and in vivo GalNAc–siRNA-mediated knockdown of mARC1 lowered hepatic but increased plasma triglycerides. We found alterations in pathways regulating lipid metabolism and decreased secretion of 3-hydroxybutyrate upon mARC1 knockdown in vitro and in vivo.
Conclusions
Collectively, our findings from human genetics, and in vitro and in vivo hepatocyte-specific mARC1 knockdown support the potential efficacy of hepatocyte-specific targeting of mARC1 for treatment of NAFLD.
Impact and implications
We report that genetically predicted increases in MTARC1 mRNA associate with poor liver health. Furthermore, knockdown of mARC1 reduces hepatic steatosis in primary human hepatocytes and a murine NASH model. Together, these findings further underscore the therapeutic potential of targeting hepatocyte MTARC1 for NAFLD.
Graphical abstract
Highlights
Related collections
Most cited references33

- Record: found
- Abstract: found
- Article: found
The UK Biobank resource with deep phenotyping and genomic data

- Record: found
- Abstract: found
- Article: found
MetaboLights: a resource evolving in response to the needs of its scientific community

- Record: found
- Abstract: found
- Article: found