- Record: found
- Abstract: found
- Article: found
SkipCPP-Pred: an improved and promising sequence-based predictor for predicting cell-penetrating peptides

Read this article at
Abstract
Background
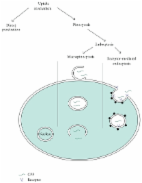
Cell-penetrating peptides (CPPs) are short peptides (5–30 amino acids) that can enter almost any cell without significant damage. On account of their high delivery efficiency, CPPs are promising candidates for gene therapy and cancer treatment. Accordingly, techniques that correctly predict CPPs are anticipated to accelerate CPP applications in future therapeutics. Recently, computational methods have been reportedly successful in predicting CPPs. Unfortunately, the predictive performance of existing methods is not satisfactory and reliable so as to accurately identify CPPs.
Results
In this study, we propose a novel computational predictor called SkipCPP-Pred to further improve the predictive performance. The novelty of the proposed predictor is that we present a sequence-based feature representation algorithm called adaptive k-skip-n-gram that sufficiently captures the intrinsic correlation information of residues. By fusing the proposed adaptive skip features with a random forest (RF) classifier, we successfully construct the prediction model of SkipCPP-Pred. The various jackknife results demonstrate that the proposed SkipCPP-Pred is 3.6% higher than state-of-the-art CPP predictors in terms of accuracy. Moreover, we construct a high-quality benchmark dataset by reducing the data redundancy and enhancing the similarity between the positive and negative classes. Using this dataset to build prediction models, we can successfully avoid the performance bias lying in existing methods and yield a promising predictive model.
Conclusions
The proposed SkipCPP-Pred is a simple and fast sequence-based predictor featured with the adaptive k-skip-n-gram model for the improved prediction of CPPs. Currently, SkipCPP-Pred is publicly available from an online webserver ( http://server.malab.cn/SkipCPP-Pred/Index.html).
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
The meaning and use of the area under a receiver operating characteristic (ROC) curve.
- Record: found
- Abstract: found
- Article: not found
Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery.
- Record: found
- Abstract: found
- Article: found