- Record: found
- Abstract: found
- Article: not found
RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview

Read this article at
Abstract
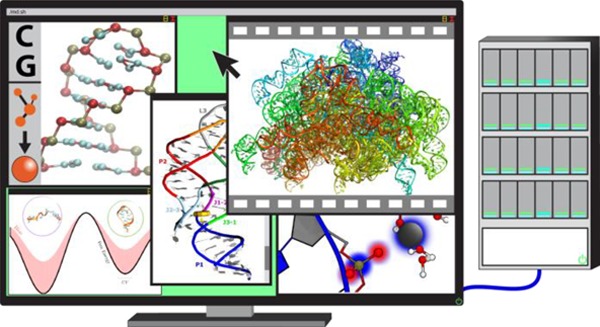
With both catalytic and genetic functions, ribonucleic acid (RNA) is perhaps the most pluripotent chemical species in molecular biology, and its functions are intimately linked to its structure and dynamics. Computer simulations, and in particular atomistic molecular dynamics (MD), allow structural dynamics of biomolecular systems to be investigated with unprecedented temporal and spatial resolution. We here provide a comprehensive overview of the fast-developing field of MD simulations of RNA molecules. We begin with an in-depth, evaluatory coverage of the most fundamental methodological challenges that set the basis for the future development of the field, in particular, the current developments and inherent physical limitations of the atomistic force fields and the recent advances in a broad spectrum of enhanced sampling methods. We also survey the closely related field of coarse-grained modeling of RNA systems. After dealing with the methodological aspects, we provide an exhaustive overview of the available RNA simulation literature, ranging from studies of the smallest RNA oligonucleotides to investigations of the entire ribosome. Our review encompasses tetranucleotides, tetraloops, a number of small RNA motifs, A-helix RNA, kissing-loop complexes, the TAR RNA element, the decoding center and other important regions of the ribosome, as well as assorted others systems. Extended sections are devoted to RNA–ion interactions, ribozymes, riboswitches, and protein/RNA complexes. Our overview is written for as broad of an audience as possible, aiming to provide a much-needed interdisciplinary bridge between computation and experiment, together with a perspective on the future of the field.
Related collections
Most cited references1,210
- Record: found
- Abstract: found
- Article: found
A nonequilibrium equality for free energy differences
- Record: found
- Abstract: not found
- Article: not found
