- Record: found
- Abstract: found
- Article: found
A statistical framework for analyzing deep mutational scanning data
Read this article at
Abstract
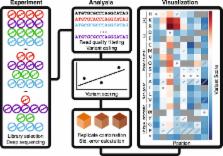
Deep mutational scanning is a widely used method for multiplex measurement of functional consequences of protein variants. We developed a new deep mutational scanning statistical model that generates error estimates for each measurement, capturing both sampling error and consistency between replicates. We apply our model to one novel and five published datasets comprising 243,732 variants and demonstrate its superiority in removing noisy variants and conducting hypothesis testing. Simulations show our model applies to scans based on cell growth or binding and handles common experimental errors. We implemented our model in Enrich2, software that can empower researchers analyzing deep mutational scanning data.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Article: not found
A general method applicable to the search for similarities in the amino acid sequence of two proteins
- Record: found
- Abstract: not found
- Article: not found