- Record: found
- Abstract: found
- Article: found
Novel symbionts and potential human pathogens excavated from argasid tick microbiomes that are shaped by dual or single symbiosis

Read this article at
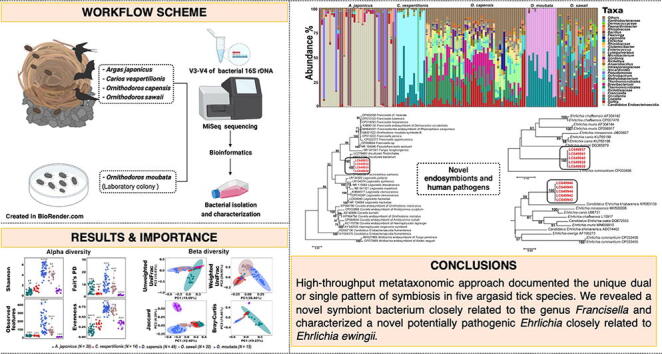
Graphical abstract
Abstract
Research on vector-associated microbiomes has been expanding due to increasing emergence of vector-borne pathogens and awareness of the importance of symbionts in the vector physiology. However, little is known about microbiomes of argasid (or soft-bodied) ticks due to limited access to specimens. We collected four argasid species ( Argas japonicus, Carios vespertilionis, Ornithodoros capensis, and Ornithodoros sawaii) from the nests or burrows of their vertebrate hosts. One laboratory-reared argasid species ( Ornithodoros moubata) was also included. Attempts were then made to isolate and characterize potential symbionts/pathogens using arthropod cell lines. Microbial community structure was distinct for each tick species. Coxiella was detected as the predominant symbiont in four tick species where dual symbiosis between Coxiella and Rickettsia or Coxiella and Francisella was observed in C. vespertilionis and O. moubata, respectively. Of note, A. japonicus lacked Coxiella and instead had Occidentia massiliensis and Thiotrichales as alternative symbionts. Our study found strong correlation between tick species and life stage. We successfully isolated Oc. massiliensis and characterized potential pathogens of genera Ehrlichia and Borrelia. The results suggest that there is no consistent trend of microbiomes in relation to tick life stage that fit all tick species and that the final interpretation should be related to the balance between environmental bacterial exposure and endosymbiont ecology. Nevertheless, our findings provide insights on the ecology of tick microbiomes and basis for future investigations on the capacity of argasid ticks to carry novel pathogens with public health importance.
Related collections
Most cited references129

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.
- Record: found
- Abstract: found
- Article: not found