- Record: found
- Abstract: found
- Article: not found
A structural analysis of M protein in coronavirus assembly and morphology
Read this article at
Abstract
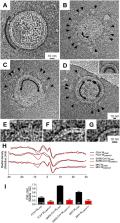
The M protein of coronavirus plays a central role in virus assembly, turning cellular membranes into workshops where virus and host factors come together to make new virus particles. We investigated how M structure and organization is related to virus shape and size using cryo-electron microscopy, tomography and statistical analysis. We present evidence that suggests M can adopt two conformations and that membrane curvature is regulated by one M conformer. Elongated M protein is associated with rigidity, clusters of spikes and a relatively narrow range of membrane curvature. In contrast, compact M protein is associated with flexibility and low spike density. Analysis of several types of virus-like particles and virions revealed that S protein, N protein and genomic RNA each help to regulate virion size and variation, presumably through interactions with M. These findings provide insight into how M protein functions to promote virus assembly.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy.
- Record: found
- Abstract: found
- Article: not found
Influenza virus pleiomorphy characterized by cryoelectron tomography.
- Record: found
- Abstract: found
- Article: not found
