- Record: found
- Abstract: found
- Article: found
Analysis of Bovine Viral Diarrhea Viruses-infected monocytes: identification of cytopathic and non-cytopathic biotype differences
Read this article at
Abstract
Background
Bovine Viral Diarrhea Virus (BVDV) infection is widespread in cattle worldwide, causing important economic losses. Pathogenesis of the disease caused by BVDV is complex, as each BVDV strain has two biotypes: non-cytopathic (ncp) and cytopathic (cp). BVDV can cause a persistent latent infection and immune suppression if animals are infected with an ncp biotype during early gestation, followed by a subsequent infection of the cp biotype. The molecular mechanisms that underscore the complex disease etiology leading to immune suppression in cattle caused by BVDV are not well understood.
Results
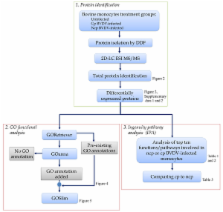
Using proteomics, we evaluated the effect of cp and ncp BVDV infection of bovine monocytes to determine their role in viral immune suppression and uncontrolled inflammation. Proteins were isolated by differential detergent fractionation and identified by 2D-LC ESI MS/MS. We identified 137 and 228 significantly altered bovine proteins due to ncp and cp BVDV infection, respectively. Functional analysis of these proteins using the Gene Ontology (GO) showed multiple under- and over- represented GO functions in molecular function, biological process and cellular component between the two BVDV biotypes. Analysis of the top immunological pathways affected by BVDV infection revealed that pathways representing macropinocytosis signalling, virus entry via endocytic pathway, integrin signalling and primary immunodeficiency signalling were identified only in ncp BVDV-infected monocytes. In contrast, pathways like actin cytoskeleton signalling, RhoA signalling, clathrin-mediated endocytosis signalling and interferon signalling were identified only in cp BDVD-infected cells. Of the six common pathways involved in cp and ncp BVDV infection, acute phase response signalling was the most significant for both BVDV biotypes. Although, most shared altered host proteins between both BVDV biotypes showed the same type of change, integrin alpha 2b (ITGA2B) and integrin beta 3 (ITGB3) were down- regulated by ncp BVDV and up- regulated by cp BVDV infection.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Regulation of actin assembly associated with protrusion and adhesion in cell migration.
- Record: found
- Abstract: found
- Article: not found
Viral infection and iron metabolism.

- Record: found
- Abstract: found
- Article: found