- Record: found
- Abstract: found
- Article: found
Study of FoxA Pioneer Factor at Silent Genes Reveals Rfx-Repressed Enhancer at Cdx2 and a Potential Indicator of Esophageal Adenocarcinoma Development
Read this article at
Abstract
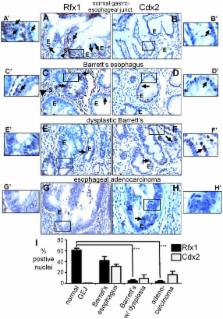
Understanding how silent genes can be competent for activation provides insight into development as well as cellular reprogramming and pathogenesis. We performed genomic location analysis of the pioneer transcription factor FoxA in the adult mouse liver and found that about one-third of the FoxA bound sites are near silent genes, including genes without detectable RNA polymerase II. Virtually all of the FoxA-bound silent sites are within conserved sequences, suggesting possible function. Such sites are enriched in motifs for transcriptional repressors, including for Rfx1 and type II nuclear hormone receptors. We found one such target site at a cryptic “shadow” enhancer 7 kilobases (kb) downstream of the Cdx2 gene, where Rfx1 restricts transcriptional activation by FoxA. The Cdx2 shadow enhancer exhibits a subset of regulatory properties of the upstream Cdx2 promoter region. While Cdx2 is ectopically induced in the early metaplastic condition of Barrett's esophagus, its expression is not necessarily present in progressive Barrett's with dysplasia or adenocarcinoma. By contrast, we find that Rfx1 expression in the esophageal epithelium becomes gradually extinguished during progression to cancer, i.e, expression of Rfx1 decreased markedly in dysplasia and adenocarcinoma. We propose that this decreased expression of Rfx1 could be an indicator of progression from Barrett's esophagus to adenocarcinoma and that similar analyses of other transcription factors bound to silent genes can reveal unanticipated regulatory insights into oncogenic progression and cellular reprogramming.
Author Summary
FoxA transcriptional regulatory proteins are “pioneer factors” that engage silent genes, helping to endow the competence for activation. About a third of the DNA sites we found to be occupied by FoxA in the adult liver are at genes that are silent. Analysis of transcription factor binding motifs near the FoxA sites at silent genes revealed a co-occurrence of motifs for the transcriptional repressors Rfx1 and type II nuclear hormone receptors (NHR-II). Further analysis of one such region downstream of the Cdx2 gene shows that it is a cryptic enhancer, in that it functions poorly unless Rfx1 or NHR-II binding is prevented, in which case FoxA1 promotes enhancer activity. Cdx2 encodes a transcription factor that promotes intestinal differentiation; ectopic expression of Cdx2 in the esophagus can help promote metaplasia and cancer. By screening numerous staged samples of human tissues, we show that Rfx1 expression is extinguished during the progression to esophageal adenocarcinoma and thus may serve as a marker of cancer progression. These studies exemplify how the analysis of pioneer factors bound to silent genes can reveal a basis for the competence of cells to deregulate gene expression and undergo transitions to cancer.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
JASPAR: an open-access database for eukaryotic transcription factor binding profiles.
- Record: found
- Abstract: found
- Article: not found
Endogenous bile acids are ligands for the nuclear receptor FXR/BAR.
- Record: found
- Abstract: found
- Article: not found
Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.