- Record: found
- Abstract: found
- Article: found
From Pioneer to Repressor: Bimodal foxd3 Activity Dynamically Remodels Neural Crest Regulatory Landscape In Vivo
Read this article at
Summary
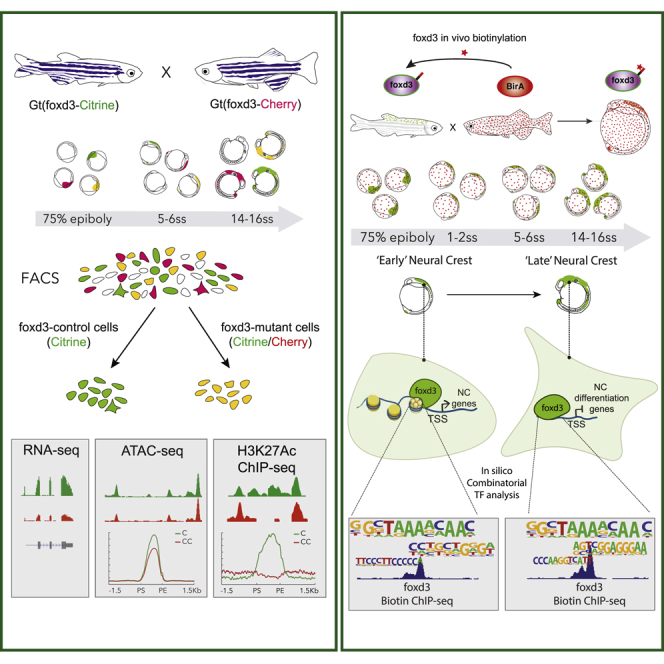
The neural crest (NC) is a transient embryonic stem cell-like population characterized by its multipotency and broad developmental potential. Here, we perform NC-specific transcriptional and epigenomic profiling of foxd3-mutant cells in vivo to define the gene regulatory circuits controlling NC specification. Together with global binding analysis obtained by foxd3 biotin-ChIP and single cell profiles of foxd3-expressing premigratory NC, our analysis shows that, during early steps of NC formation, foxd3 acts globally as a pioneer factor to prime the onset of genes regulating NC specification and migration by re-arranging the chromatin landscape, opening cis-regulatory elements and reshuffling nucleosomes. Strikingly, foxd3 then gradually switches from an activator to its well-described role as a transcriptional repressor and potentially uses differential partners for each role. Taken together, these results demonstrate that foxd3 acts bimodally in the neural crest as a switch from “permissive” to “repressive” nucleosome and chromatin organization to maintain multipotency and define cell fates.
Graphical Abstract
Highlights
-
•
FoxD3 primes neural crest specification by modulating distal enhancers
-
•
FoxD3 represses a number of neural crest migration and differentiation genes
-
•
In neural crest, FoxD3 acts to switch chromatin from “permissive” to “repressive”
-
•
Distinctive gene regulatory mechanisms underlie the bimodal action of FoxD3
Abstract
Through transcriptional and epigenomic profiling of foxd3-mutant zebrafish neural crest cells (NCCs) and whole-genome mapping of FoxD3 binding, Lukoseviciute et al. uncover bimodal FoxD3 action across NCC development. FoxD3 acts as a pioneer factor to prime genes for NCC specification before switching to being a repressor to control migration and differentiation.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
Transcription factors: from enhancer binding to developmental control.

- Record: found
- Abstract: found
- Article: found
seqMINER: an integrated ChIP-seq data interpretation platform
- Record: found
- Abstract: found
- Article: not found
The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.