- Record: found
- Abstract: found
- Article: found
A New Approach of Fatigue Classification Based on Data of Tongue and Pulse With Machine Learning
Read this article at
Abstract
Background
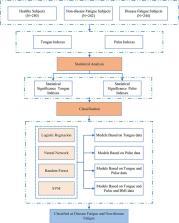
Fatigue is a common and subjective symptom, which is associated with many diseases and suboptimal health status. A reliable and evidence-based approach is lacking to distinguish disease fatigue and non-disease fatigue. This study aimed to establish a method for early differential diagnosis of fatigue, which can be used to distinguish disease fatigue from non-disease fatigue, and to investigate the feasibility of characterizing fatigue states in a view of tongue and pulse data analysis.
Methods
Tongue and Face Diagnosis Analysis-1 (TFDA-1) instrument and Pulse Diagnosis Analysis-1 (PDA-1) instrument were used to collect tongue and pulse data. Four machine learning models were used to perform classification experiments of disease fatigue vs. non-disease fatigue.
Results
The results showed that all the four classifiers over “Tongue & Pulse” joint data showed better performances than those only over tongue data or only over pulse data. The model accuracy rates based on logistic regression, support vector machine, random forest, and neural network were (85.51 ± 1.87)%, (83.78 ± 4.39)%, (83.27 ± 3.48)% and (85.82 ± 3.01)%, and with Area Under Curve estimates of 0.9160 ± 0.0136, 0.9106 ± 0.0365, 0.8959 ± 0.0254 and 0.9239 ± 0.0174, respectively.
Related collections
Most cited references49

- Record: found
- Abstract: found
- Article: found
Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning
- Record: found
- Abstract: found
- Article: not found
eDoctor: machine learning and the future of medicine.
- Record: found
- Abstract: found
- Article: not found