- Record: found
- Abstract: found
- Article: found
Core circadian clock and light signaling genes brought into genetic linkage across the green lineage
Read this article at
Abstract
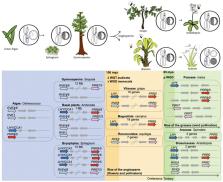
The circadian clock is conserved at both the level of transcriptional networks as well as core genes in plants, ensuring that biological processes are phased to the correct time of day. In the model plant Arabidopsis ( Arabidopsis thaliana), the core circadian SHAQKYF-type-MYB ( sMYB) genes CIRCADIAN CLOCK ASSOCIATED 1 ( CCA1) and REVEILLE ( RVE4) show genetic linkage with PSEUDO-RESPONSE REGULATOR 9 ( PRR9) and PRR7, respectively. Leveraging chromosome-resolved plant genomes and syntenic ortholog analysis enabled tracing this genetic linkage back to Amborella trichopoda, a sister lineage to the angiosperm, and identifying an additional evolutionarily conserved genetic linkage in light signaling genes. The LHY/CCA1–PRR5/9, RVE4/8–PRR3/7, and PIF3–PHYA genetic linkages emerged in the bryophyte lineage and progressively moved within several genes of each other across an array of angiosperm families representing distinct whole-genome duplication and fractionation events. Soybean ( Glycine max) maintained all but two genetic linkages, and expression analysis revealed the PIF3–PHYA linkage overlapping with the E4 maturity group locus was the only pair to robustly cycle with an evening phase, in contrast to the sMYB–PRR morning and midday phase. While most monocots maintain the genetic linkages, they have been lost in the economically important grasses (Poaceae), such as maize ( Zea mays), where the genes have been fractionated to separate chromosomes and presence/absence variation results in the segregation of PRR7 paralogs across heterotic groups. The environmental robustness model is put forward, suggesting that evolutionarily conserved genetic linkages ensure superior microhabitat pollinator synchrony, while wide-hybrids or unlinking the genes, as seen in the grasses, result in heterosis, adaptation, and colonization of new ecological niches.
Abstract
The genetic linkage of the core circadian clock and light signaling genes coincides with the rise to dominance of flowering plants and may explain environment-specific growth as well as heterosis.
Related collections
Most cited references148

- Record: found
- Abstract: found
- Article: found
OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy

- Record: found
- Abstract: found
- Article: found
One thousand plant transcriptomes and the phylogenomics of green plants
- Record: found
- Abstract: found
- Article: not found