- Record: found
- Abstract: found
- Article: found
Computational analysis of sense-antisense chimeric transcripts reveals their potential regulatory features and the landscape of expression in human cells
Read this article at
Abstract
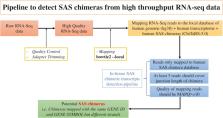
Many human genes are transcribed from both strands and produce sense-antisense gene pairs. Sense-antisense (SAS) chimeric transcripts are produced upon the coalescing of exons/introns from both sense and antisense transcripts of the same gene. SAS chimera was first reported in prostate cancer cells. Subsequently, numerous SAS chimeras have been reported in the ChiTaRS-2.1 database. However, the landscape of their expression in human cells and functional aspects are still unknown. We found that longer palindromic sequences are a unique feature of SAS chimeras. Structural analysis indicates that a long hairpin-like structure formed by many consecutive Watson-Crick base pairs appears because of these long palindromic sequences, which possibly play a similar role as double-stranded RNA (dsRNA), interfering with gene expression. RNA–RNA interaction analysis suggested that SAS chimeras could significantly interact with their parental mRNAs, indicating their potential regulatory features. Here, 267 SAS chimeras were mapped in RNA-seq data from 16 healthy human tissues, revealing their expression in normal cells. Evolutionary analysis suggested the positive selection favoring sense-antisense fusions that significantly impacted the evolution of their function and structure. Overall, our study provides detailed insight into the expression landscape of SAS chimeras in human cells and identifies potential regulatory features.
Related collections
Most cited references110
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found