- Record: found
- Abstract: found
- Article: found
Discovery of the fourth mobile sulfonamide resistance gene
Read this article at
Abstract
Background
Over the past 75 years, human pathogens have acquired antibiotic resistance genes (ARGs), often from environmental bacteria. Integrons play a major role in the acquisition of antibiotic resistance genes. We therefore hypothesized that focused exploration of integron gene cassettes from microbial communities could be an efficient way to find novel mobile resistance genes. DNA from polluted Indian river sediments were amplified using three sets of primers targeting class 1 integrons and sequenced by long- and short-read technologies to maintain both accuracy and context.
Results
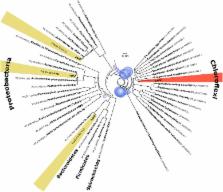
Up to 89% of identified open reading frames encode known resistance genes, or variations thereof (> 1000). We identified putative novel ARGs to aminoglycosides, beta-lactams, trimethoprim, rifampicin, and chloramphenicol, including several novel OXA variants, providing reduced susceptibility to carbapenems. One dihydropteroate synthase gene, with less than 34% amino acid identity to the three known mobile sulfonamide resistance genes ( sul1–3), provided complete resistance when expressed in Escherichia coli. The mobilized gene, here named sul4, is the first mobile sulfonamide resistance gene discovered since 2003. Analyses of adjacent DNA suggest that sul4 has been decontextualized from a set of chromosomal genes involved in folate synthesis in its original host, likely within the phylum Chloroflexi. The presence of an insertion sequence common region element could provide mobility to the entire integron. Screening of 6489 metagenomic datasets revealed that sul4 is already widespread in seven countries across Asia and Europe.
Conclusions
Our findings show that exploring integrons from environmental communities with a history of antibiotic exposure can provide an efficient way to find novel, mobile resistance genes. The mobilization of a fourth sulfonamide resistance gene is likely to provide expanded opportunities for sulfonamide resistance to spread, with potential impacts on both human and animal health.
Related collections
Most cited references49

- Record: found
- Abstract: found
- Article: found
ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data
- Record: found
- Abstract: found
- Article: not found
The comprehensive antibiotic resistance database.
- Record: found
- Abstract: not found
- Book: not found