- Record: found
- Abstract: found
- Article: found
Gut Mucosal Proteins and Bacteriome Are Shaped by the Saturation Index of Dietary Lipids

Read this article at
Abstract
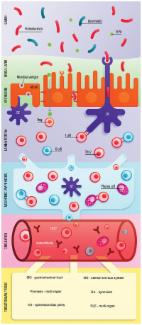
The dynamics of the tripartite relationship between the host, gut bacteria and diet in the gut is relatively unknown. An imbalance between harmful and protective gut bacteria, termed dysbiosis, has been linked to many diseases and has most often been attributed to high-fat dietary intake. However, we recently clarified that the type of fat, not calories, were important in the development of murine colitis. To further understand the host-microbe dynamic in response to dietary lipids, we fed mice isocaloric high-fat diets containing either milk fat, corn oil or olive oil and performed 16S rRNA gene sequencing of the colon microbiome and mass spectrometry-based relative quantification of the colonic metaproteome. The corn oil diet, rich in omega-6 polyunsaturated fatty acids, increased the potential for pathobiont survival and invasion in an inflamed, oxidized and damaged gut while saturated fatty acids promoted compensatory inflammatory responses involved in tissue healing. We conclude that various lipids uniquely alter the host-microbe interaction in the gut. While high-fat consumption has a distinct impact on the gut microbiota, the type of fatty acids alters the relative microbial abundances and predicted functions. These results support that the type of fat are key to understanding the biological effects of high-fat diets on gut health.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
The core gut microbiome, energy balance and obesity.
- Record: found
- Abstract: found
- Article: found
The Gut Microbiota in Immune-Mediated Inflammatory Diseases
- Record: found
- Abstract: found
- Article: not found