- Record: found
- Abstract: found
- Article: found
Predominately Uncultured Microbes as Sources of Bioactive Agents
Read this article at
Abstract
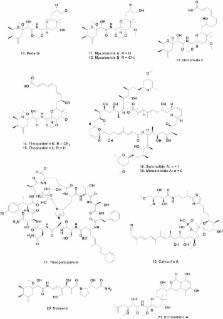
In this short review, I am discussing the relatively recent awareness of the role of symbionts in plant, marine-invertebrates and fungal areas. It is now quite obvious that in marine-invertebrates, a majority of compounds found are from either as yet unculturable or poorly culturable microbes, and techniques involving “state of the art” genomic analyses and subsequent computerized analyses are required to investigate these interactions. In the plant kingdom evidence is amassing that endophytes (mainly fungal in nature) are heavily involved in secondary metabolite production and that mimicking the microbial interactions of fermentable microbes leads to involvement of previously unrecognized gene clusters (cryptic clusters is one name used), that when activated, produce previously unknown bioactive molecules.
Related collections
Most cited references120
- Record: found
- Abstract: found
- Article: not found
Discovery of microbial natural products by activation of silent biosynthetic gene clusters.
- Record: found
- Abstract: found
- Article: not found
The evolution of genome mining in microbes - a review.
- Record: found
- Abstract: found
- Article: not found