- Record: found
- Abstract: found
- Article: found
Multidrug-resistant and enterotoxigenic methicillin-resistant Staphylococcus aureus isolated from raw milk of cows at small-scale production units
Read this article at
Abstract
Objective:
Staphylococcus aureus ( S. aureus) has evolved as one of the most significant bacteria causing food poisoning outbreaks worldwide. This study was carried out to investigate the prevalence, antibiotic sensitivity, virulence, and enterotoxin production of S. aureus in raw milk of cow from small-scale production units and house-raised animals in Damietta governorate, Egypt.
Material and Methods:
The samples were examined bacteriologically, and antimicrobial sensitivity testing was carried out. Moreover, isolates were characterized by the molecular detection of antimicrobial resistance, virulence, and enterotoxin genes.
Results:
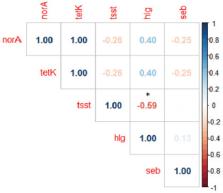
Out of 300 milk samples examined, S. aureus was isolated from 50 samples (16.7%). Antibiotic sensitivity testing revealed that isolates were resistant to β-lactams (32%), tetracycline (16%), and norfloxacin (16%); however, they showed considerable sensitivity to ceftaroline and amikacin (72%). Multidrug-resistance (MDR) has been observed in eight isolates (16%), with a MDR index (0.5) in all of them. Of the total S. aureus isolates obtained, methicillin-resistant S. aureus (MRSA) has been confirmed molecularly in 16/50 (32%) and was found to carry mecA and coa genes, while virulence genes; hlg (11/16, 68.75%) and tsst (6/16, 37.5%) were amplified at a lower percentage, and they showed a significant moderate negative correlation ( r = −0.59, p-value > 0.05). Antibiotic resistance genes have been detected in resistant isolates relevant to their phenotypic resistance: blaZ (100%), tetK (50%), and norA (50%). Fifty percent of MRSA isolates carried the seb enterotoxin gene.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences
- Record: found
- Abstract: found
- Article: not found
