- Record: found
- Abstract: found
- Article: found
Expressed sequence tags (ESTs) from immune tissues of turbot ( Scophthalmus maximus) challenged with pathogens
Read this article at
Abstract
Background
The turbot ( Scophthalmus maximus; Scophthalmidae; Pleuronectiformes) is a flatfish species of great relevance for marine aquaculture in Europe. In contrast to other cultured flatfish, very few genomic resources are available in this species. Aeromonas salmonicida and Philasterides dicentrarchi are two pathogens that affect turbot culture causing serious economic losses to the turbot industry. Little is known about the molecular mechanisms for disease resistance and host-pathogen interactions in this species. In this work, thousands of ESTs for functional genomic studies and potential markers linked to ESTs for mapping (microsatellites and single nucleotide polymorphisms (SNPs)) are provided. This information enabled us to obtain a preliminary view of regulated genes in response to these pathogens and it constitutes the basis for subsequent and more accurate microarray analysis.
Results
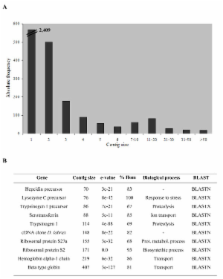
A total of 12584 cDNAs partially sequenced from three different cDNA libraries of turbot ( Scophthalmus maximus) infected with Aeromonas salmonicida, Philasterides dicentrarchi and from healthy fish were analyzed. Three immune-relevant tissues (liver, spleen and head kidney) were sampled at several time points in the infection process for library construction. The sequences were processed into 9256 high-quality sequences, which constituted the source for the turbot EST database. Clustering and assembly of these sequences, revealed 3482 different putative transcripts, 1073 contigs and 2409 singletons. BLAST searches with public databases detected significant similarity (e-value ≤ 1e-5) in 1766 (50.7%) sequences and 816 of them (23.4%) could be functionally annotated. Two hundred three of these genes (24.9%), encoding for defence/immune-related proteins, were mostly identified for the first time in turbot. Some ESTs showed significant differences in the number of transcripts when comparing the three libraries, suggesting regulation in response to these pathogens. A total of 191 microsatellites, with 104 having sufficient flanking sequences for primer design, and 1158 putative SNPs were identified from these EST resources in turbot.
Conclusion
A collection of 9256 high-quality ESTs was generated representing 3482 unique turbot sequences. A large proportion of defence/immune-related genes were identified, many of them regulated in response to specific pathogens. Putative microsatellites and SNPs were identified. These genome resources constitute the basis to develop a microarray for functional genomics studies and marker validation for genetic linkage and QTL analysis in turbot.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Accessing genetic variation: genotyping single nucleotide polymorphisms.
- Record: found
- Abstract: not found
- Article: not found