- Record: found
- Abstract: found
- Article: found
Requirements for data integration platforms in biomedical research networks: a reference model
Read this article at
Abstract
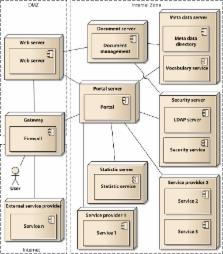
Biomedical research networks need to integrate research data among their members and with external partners. To support such data sharing activities, an adequate information technology infrastructure is necessary. To facilitate the establishment of such an infrastructure, we developed a reference model for the requirements. The reference model consists of five reference goals and 15 reference requirements. Using the Unified Modeling Language, the goals and requirements are set into relation to each other. In addition, all goals and requirements are described textually in tables. This reference model can be used by research networks as a basis for a resource efficient acquisition of their project specific requirements. Furthermore, a concrete instance of the reference model is described for a research network on liver cancer. The reference model is transferred into a requirements model of the specific network. Based on this concrete requirements model, a service-oriented information technology architecture is derived and also described in this paper.
Related collections
Most cited references27
- Record: found
- Abstract: not found
- Book: not found
R: A language and environment for statistical computing
- Record: found
- Abstract: found
- Article: not found
Desiderata for controlled medical vocabularies in the twenty-first century.
- Record: found
- Abstract: not found
- Article: not found