- Record: found
- Abstract: found
- Article: found
Characterization of cancer-related fibroblasts (CAF) in hepatocellular carcinoma and construction of CAF-based risk signature based on single-cell RNA-seq and bulk RNA-seq data
Read this article at
Abstract
Background
Cancer-associated fibroblasts (CAFs) are involved in tumor growth, angiogenesis, metastasis, and resistance to therapy. We sought to explore the CAFs characteristics in hepatocellular carcinoma (HCC) and establish a CAF-based risk signature for predicting the prognosis of HCC patients.
Methods
The signal-cell RNA sequencing (scRNA-seq) data was obtained from the GEO database. Bulk RNA-seq data and microarray data of HCC were obtained from the TCGA and GEO databases respectively. Seurat R package was applied to process scRNA-seq data and identify CAF clusters according to the CAF markers. Differential expression analysis was performed to screen differentially expressed genes (DEGs) between normal and tumor samples in TCGA dataset. Then Pearson correlation analysis was used to determine the DEGs associated with CAF clusters, followed by the univariate Cox regression analysis to identify CAF-related prognostic genes. Lasso regression was implemented to construct a risk signature based on CAF-related prognostic genes. Finally, a nomogram model based on the risk signature and clinicopathological characteristics was developed.
Results
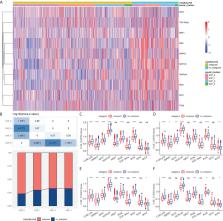
Based on scRNA-seq data, we identified 4 CAF clusters in HCC, 3 of which were associated with prognosis in HCC. A total of 423 genes were identified from 2811 DEGs to be significantly correlated with CAF clusters, and were narrowed down to generate a risk signature with 6 genes. These six genes were primarily connected with 39 pathways, such as angiogenesis, apoptosis, and hypoxia. Meanwhile, the risk signature was significantly associated with stromal and immune scores, as well as some immune cells. Multivariate analysis revealed that risk signature was an independent prognostic factor for HCC, and its value in predicting immunotherapeutic outcomes was confirmed. A novel nomogram integrating the stage and CAF-based risk signature was constructed, which exhibited favorable predictability and reliability in the prognosis prediction of HCC.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found