- Record: found
- Abstract: found
- Article: found
Strong Selectional Forces Fine-Tune CpG Content in Genes Involved in Neurological Disorders as Revealed by Codon Usage Patterns
Read this article at
Abstract
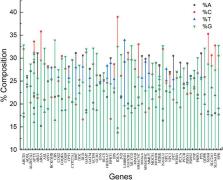
Neurodegenerative disorders cause irreversible damage to the neurons and adversely affect the quality of life. Protein misfolding and their aggregation in specific parts of the brain, mitochondrial dysfunction, calcium load, proteolytic stress, and oxidative stress are among the causes of neurodegenerative disorders. In addition, altered metabolism has been associated with neurodegeneration as evidenced by reductions in glutamine and alanine in transient global amnesia patients, higher homocysteine-cysteine disulfide, and lower methionine decline in serum urea have been observed in Alzheimer’s disease patients. Neurodegeneration thus appears to be a culmination of altered metabolism. The study’s objective is to analyze various attributes like composition, physical properties of the protein, and factors like selectional and mutational forces, influencing codon usage preferences in a panel of genes involved directly or indirectly in metabolism and contributing to neurodegeneration. Various parameters, including gene composition, dinucleotide analysis, Relative synonymous codon usage (RSCU), Codon adaptation index (CAI), neutrality and parity plots, and different protein indices, were computed and analyzed to determine the codon usage pattern and factors affecting it. The correlation of intrinsic protein properties such as the grand average of hydropathicity index (GRAVY), isoelectric point, hydrophobicity, and acidic, basic, and neutral amino acid content has been found to influence codon usage. In genes up to 800 amino acids long, the GC3 content was highly variable, while GC12 content was relatively constant. An optimum CpG content is present in genes to maintain a high expression level as required for genes involved in metabolism. Also observed was a low codon usage bias with a higher protein expression level. Compositional parameters and nucleotides at the second position of codons played essential roles in explaining the extent of bias. Overall analysis indicated that the dominance of selection pressure and compositional constraints and mutational forces shape codon usage.
Related collections
Most cited references58
- Record: found
- Abstract: not found
- Book Chapter: not found
Protein Identification and Analysis Tools on the ExPASy Server
- Record: found
- Abstract: not found
- Article: not found
A simple method for displaying the hydropathic character of a protein.
- Record: found
- Abstract: found
- Article: not found