- Record: found
- Abstract: found
- Article: found
Prime editor‐mediated functional reshaping of ACE2 prevents the entry of multiple human coronaviruses, including SARS‐CoV‐2 variants

Read this article at
Abstract
The spike protein of SARS‐CoV‐2 hijacks the host angiotensin converting enzyme 2 (ACE2) to meditate its entry and is the primary target for vaccine development. Nevertheless, SARS‐CoV‐2 keeps evolving and the latest Omicron subvariants BQ.1 and XBB have gained exceptional immune evasion potential through mutations in their spike proteins, leading to sharply reduced efficacy of current spike‐focused vaccines and therapeutics. Compared with the fast‐evolving spike protein, targeting host ACE2 offers an alternative antiviral strategy that is more resistant to viral evolution and can even provide broad prevention against SARS‐CoV and HCoV‐NL63. Here, we use prime editor (PE) to precisely edit ACE2 at structurally selected sites. We demonstrated that residue changes at Q24/D30/K31 and/or K353 of ACE2 could completely ablate the binding of tested viruses while maintaining its physiological role in host angiotensin II conversion. PE‐mediated ACE2 editing at these sites suppressed the entry of pseudotyped SARS‐CoV‐2 major variants of concern and even SARS‐CoV or HCoV‐NL63. Moreover, it significantly inhibited the replication of the Delta variant live virus. Our work investigated the unexplored application potential of prime editing in high‐risk infectious disease control and demonstrated that such gene editing‐based host factor reshaping strategy can provide broad‐spectrum antiviral activity and a high barrier to viral escape or resistance.
Abstract
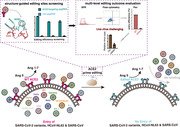
ACE2‐editing provided protection against multiple human coronaviruses (HCoVs), including SARS‐CoV, HCoV‐NL63, SARS‐CoV‐2, and its variants of concern. Model for ACE2‐editing‐provided protection against multiple HCoVs. Left: Wild‐type angiotensin converting enzyme 2 (ACE2) catalyzes the conversion of Ang II into Ang 1–7 and serves as the entry receptor for multiple HCoVs. Middle: ACE2‐editing site screening is guided by structural analysis, and editing outcomes are evaluated at multiple levels. Right: Precise editing of ACE2 blocks the cell entry of multiple HCoVs and meanwhile maintains the physiological function of ACE2 in renin–angiotensin system.
Related collections
Most cited references50

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin

- Record: found
- Abstract: found
- Article: found
SWISS-MODEL: homology modelling of protein structures and complexes
- Record: found
- Abstract: found
- Article: not found