- Record: found
- Abstract: found
- Article: found
Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma
Read this article at
Abstract
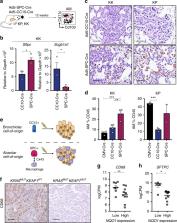
The KRAS oncoprotein, a critical driver in 33% of lung adenocarcinoma (LUAD), has remained an elusive clinical target due to its perceived undruggable nature. The identification of dependencies borne through common co-occurring mutations are sought to more effectively target KRAS-mutant lung cancer. Approximately 20% of KRAS-mutant LUAD carry loss-of-function mutations in KEAP1, a negative regulator of the antioxidant response transcription factor NFE2L2/NRF2. We demonstrate that Keap1-deficient Kras G12D lung tumors arise from a bronchiolar cell-of-origin, lacking pro-tumorigenic macrophages observed in tumors originating from alveolar cells. Keap1 loss activates the pentose phosphate pathway, inhibition of which, using 6-AN, abrogated tumor growth. These studies highlight alternative therapeutic approaches to specifically target this unique subset of KRAS-mutant LUAD cancers.
Abstract
Lung adenocarcinomas frequently harbour KRAS mutations, of which a subset are characterized by co-mutation of KEAP1. Here the authors show, in mice, that Kras G12D mutant tumours are metabolically distinct, with a bronchiolar cell-of-origin.
Related collections
Most cited references36
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.

- Record: found
- Abstract: found
- Article: found