- Record: found
- Abstract: found
- Article: found
Experimental temperatures shape host microbiome diversity and composition

Read this article at
Abstract
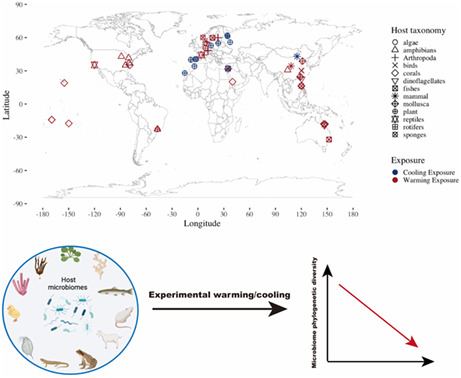
Global climate change has led to more extreme thermal events. Plants and animals harbour diverse microbial communities, which may be vital for their physiological performance and help them survive stressful climatic conditions. The extent to which microbiome communities change in response to warming or cooling may be important for predicting host performance under global change. Using a meta‐analysis of 1377 microbiomes from 43 terrestrial and aquatic species, we found a decrease in the amplicon sequence variant‐level microbiome phylogenetic diversity and alteration of microbiome composition under both experimental warming and cooling. Microbiome beta dispersion was not affected by temperature changes. We showed that the host habitat and experimental factors affected microbiome diversity and composition more than host biological traits. In particular, aquatic organisms—especially in marine habitats—experienced a greater depletion in microbiome diversity under cold conditions, compared to terrestrial hosts. Exposure involving a sudden long and static temperature shift was associated with microbiome diversity loss, but this reduction was attenuated by prior‐experimental lab acclimation or when a ramped regime (i.e., warming) was used. Microbial differential abundance and co‐occurrence network analyses revealed several potential indicator bacterial classes for hosts in heated environments and on different biome levels. Overall, our findings improve our understanding on the impact of global temperature changes on animal and plant microbiome structures across a diverse range of habitats. The next step is to link these changes to measures of host fitness, as well as microbial community functions, to determine whether microbiomes can buffer some species against a more thermally variable and extreme world.
Abstract
Related collections
Most cited references125
- Record: found
- Abstract: not found
- Article: not found
Fitting Linear Mixed-Effects Models Usinglme4

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: not found
- Article: not found