- Record: found
- Abstract: found
- Article: found
Identification of Small Nucleolar RNA SNORD60 as a Potential Biomarker and Its Clinical Significance in Lung Adenocarcinoma
Read this article at
Abstract
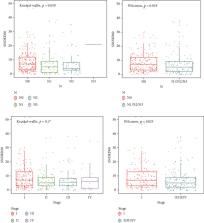
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer deaths in the world and often diagnosed at an advanced stage, so it is urgent to explore the pathogenesis and new diagnostic biomarkers. Accumulated evidences suggested that small nucleolar RNAs (snoRNAs) played a key role in the development and progression of NSCLC. To examine differential expression snoRNA profile and identify snoRNAs with clinical significance in lung adenocarcinoma (LUAD), The Cancer Genome Atlas (TCGA) LUAD RNA sequencing dataset was used to investigate differential expression snoRNA signatures and compared with snoRNA PCR array analysis in pair-matched LUAD tissues. The diagnostic ability of SONRD60 was assessed using a receiver operating characteristic (ROC) curve. The Kaplan-Meier method was used to plot survival curves. Univariate and multivariate Cox regression analyses were used to investigate the prognostic effect of SNORD60 expression on LUAD. The results showed that SNORD60 was a significantly upregulated snoRNA after intersection analysis in LUAD cases. SNORD60 has 74.2% sensitivity and 75.3% specificity for the diagnosis of LUAD. Increased SNORD60 expression was linked with lymph node metastases and the TNM stage ( P < 0.05). Pathological T category and lymph node metastases were independent prognostic factors for overall survival in a multivariate Cox regression study. Our findings demonstrated that SNORD60, a small nucleolar RNA, has an oncogenic function in LUAD and might be used as a new early diagnostic biomarker for LUAD.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
- Record: found
- Abstract: found
- Article: not found
The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification.

- Record: found
- Abstract: found
- Article: found