- Record: found
- Abstract: found
- Article: found
Using Chromatin Accessibility to Delineate Therapeutic Subtypes in Pancreatic Cancer Patient-Derived Cell Lines

Read this article at
Summary
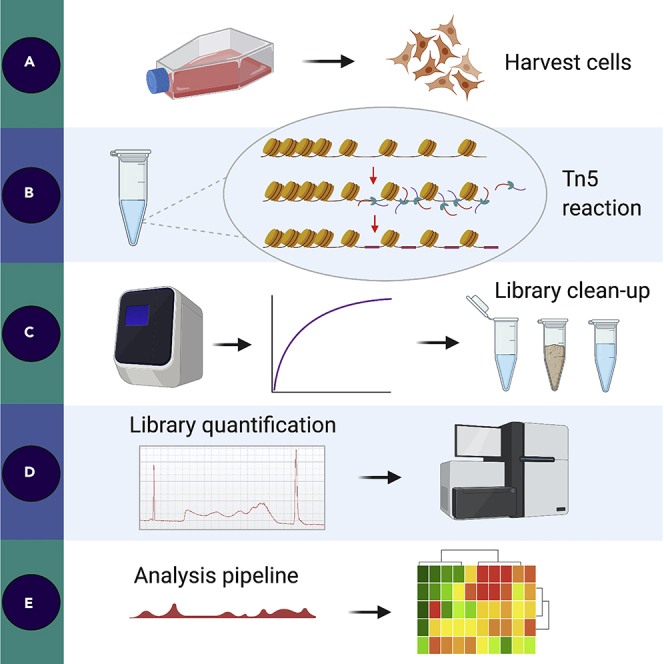
Disrupted chromatin regulatory processes contribute to the development of cancer, in particular pancreatic ductal adenocarcinoma. The assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq) is typically used to study chromatin organization. Here, we present a revised ATAC-seq protocol to study chromatin accessibility in adherent patient-derived cell lines. We provide details on how to calculate the library molarity using Agilent’s Bioanalyzer and an analysis pipeline for peak calling and transcription factor mapping.
For complete details on the use and execution of this protocol, please refer to Brunton et al. (2020).
Graphical Abstract
Highlights
Abstract
Disrupted chromatin regulatory processes contribute to the development of cancer, in particular pancreatic ductal adenocarcinoma. The assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq) is typically used to study chromatin organization. Here, we present a revised ATAC-seq protocol to study chromatin accessibility in adherent patient-derived cell lines. We provide details on how to calculate the library molarity using Agilent’s Bioanalyzer and an analysis pipeline for peak calling and transcription factor mapping.
Related collections
Most cited references6
- Record: found
- Abstract: found
- Article: not found
Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities.
- Record: found
- Abstract: found
- Article: not found
ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization.
- Record: found
- Abstract: found
- Article: not found