- Record: found
- Abstract: found
- Article: found
Coupling of Polo kinase activation to nuclear localization by a bifunctional NLS is required during mitotic entry
Read this article at
Abstract
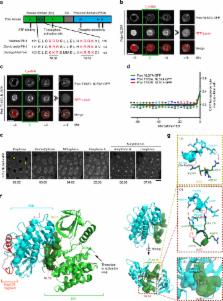
The Polo kinase is a master regulator of mitosis and cytokinesis conserved from yeasts to humans. Polo is composed of an N-term kinase domain (KD) and a C-term polo-box domain (PBD), which regulates its subcellular localizations. The PBD and KD can interact and inhibit each other, and this reciprocal inhibition is relieved when Polo is phosphorylated at its activation loop. How Polo activation and localization are coupled during mitotic entry is unknown. Here we report that PBD binding to the KD masks a nuclear localization signal (NLS). Activating phosphorylation of the KD leads to exposure of the NLS and entry of Polo into the nucleus before nuclear envelope breakdown. Failures of this mechanism result in misregulation of the Cdk1-activating Cdc25 phosphatase and lead to mitotic and developmental defects in Drosophila. These results uncover spatiotemporal mechanisms linking master regulatory enzymes during mitotic entry.
Abstract
Drosophila Polo kinase is the founding member of the Polo-Like Kinase (PLK) family and a master regulator of mitosis and cytokinesis. Here the authors uncover a molecular mechanism for the spatiotemporal regulation of Polo kinase during mitotic entry through a phosphorylation event that triggers nuclear import.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Protein kinases: evolution of dynamic regulatory proteins.
- Record: found
- Abstract: found
- Article: not found
Regulation of protein kinases; controlling activity through activation segment conformation.
- Record: found
- Abstract: found
- Article: not found