- Record: found
- Abstract: found
- Article: not found
The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development
Read this article at
Summary
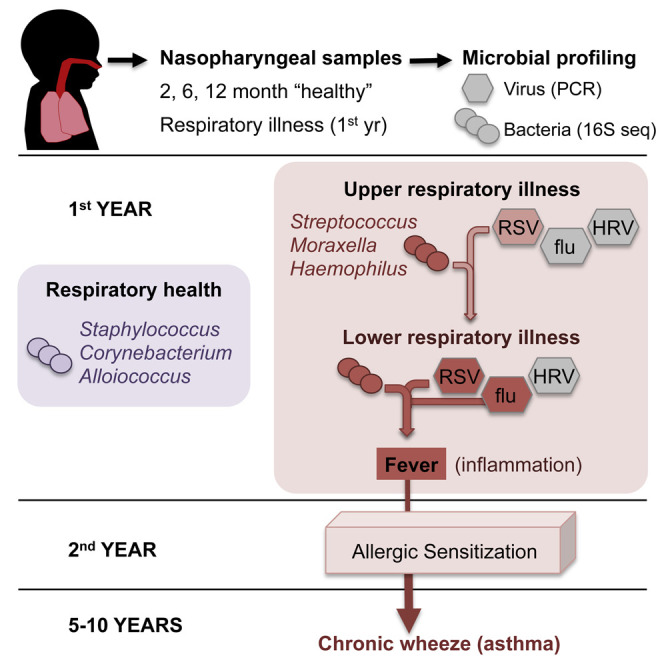
The nasopharynx (NP) is a reservoir for microbes associated with acute respiratory infections (ARIs). Lung inflammation resulting from ARIs during infancy is linked to asthma development. We examined the NP microbiome during the critical first year of life in a prospective cohort of 234 children, capturing both the viral and bacterial communities and documenting all incidents of ARIs. Most infants were initially colonized with Staphylococcus or Corynebacterium before stable colonization with Alloiococcus or Moraxella. Transient incursions of Streptococcus, Moraxella, or Haemophilus marked virus-associated ARIs. Our data identify the NP microbiome as a determinant for infection spread to the lower airways, severity of accompanying inflammatory symptoms, and risk for future asthma development. Early asymptomatic colonization with Streptococcus was a strong asthma predictor, and antibiotic usage disrupted asymptomatic colonization patterns. In the absence of effective anti-viral therapies, targeting pathogenic bacteria within the NP microbiome could represent a prophylactic approach to asthma.
Graphical Abstract
Highlights
-
•
The nasopharynx microbiome of infants has a simple structure dominated by six genera
-
•
Microbiome composition affects infection severity and pathogen spread to lower airways
-
•
Early asymptomatic colonization with Streptococcus increases risk of asthma
-
•
Antibiotic usage disrupts asymptomatic colonization patterns
Abstract
Teo et al. characterize bacterial and viral communities within the infant nasopharynx during the first year of life, comparing between asymptomatic colonization and episodes of acute respiratory infections. Microbiome composition affects infection severity and spread to lower airways and risk for future asthma development.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found
Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children.
- Record: found
- Abstract: found
- Article: not found