- Record: found
- Abstract: found
- Article: found
Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development
Read this article at
Abstract
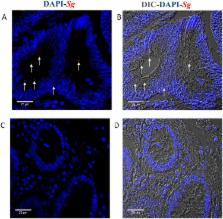
Streptococcus gallolyticus subsp. gallolyticus ( Sg) has long been known to have a strong association with colorectal cancer (CRC). This knowledge has important clinical implications, and yet little is known about the role of Sg in the development of CRC. Here we demonstrate that Sg promotes human colon cancer cell proliferation in a manner that depends on cell context, bacterial growth phase and direct contact between bacteria and colon cancer cells. In addition, we observed increased level of β-catenin, c-Myc and PCNA in colon cancer cells following incubation with Sg. Knockdown or inhibition of β-catenin abolished the effect of Sg. Furthermore, mice administered with Sg had significantly more tumors, higher tumor burden and dysplasia grade, and increased cell proliferation and β-catenin staining in colonic crypts compared to mice receiving control bacteria. Finally, we showed that Sg is present in the majority of CRC patients and is preferentially associated with tumor compared to normal tissues obtained from CRC patients. These results taken together establish for the first time a tumor-promoting role of Sg that involves specific bacterial and host factors and have important clinical implications.
Author summary
Colorectal cancer (CRC) is a leading cause of cancer-related death. The recognition that microbial agents can contribute to the development of CRC raises hope for improving CRC diagnosis and treatment by incorporating both microbial and patient characteristics into clinical strategies. S. gallolyticus subsp. gallolyticus ( Sg) has been implicated in CRC for decades. Patients with Sg infections display a much higher risk of having CRC compared to the general population. Despite this, the precise role of Sg in the development of CRC— i. e., whether this organism plays an active role in the development of tumor or its presence is merely a consequence of the tumor environment being favorable for its colonization of the colon—was unknown. Here using in vitro cell cultures and mouse models of CRC, we demonstrate that Sg actively promotes colon cancer cell proliferation and tumor growth, suggesting that it is not an innocent “passenger”. These results represent a major step forward in understanding the relationship between Sg and CRC. This combined with the prevalence of Sg in CRC patients highlight Sg as being both clinically relevant and functionally important for CRC.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Identification of c-MYC as a target of the APC pathway.
- Record: found
- Abstract: found
- Article: not found
Wnt/beta-catenin signaling in cancer stemness and malignant behavior.
- Record: found
- Abstract: found
- Article: not found