- Record: found
- Abstract: found
- Article: found
An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation
Read this article at
Abstract
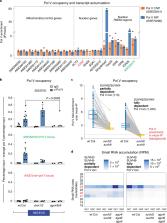
In mammals and plants, cytosine DNA methylation is essential for the epigenetic repression of transposable elements and foreign DNA. In plants, DNA methylation is guided by small interfering RNAs (siRNAs) in a self-reinforcing cycle termed RNA-directed DNA methylation (RdDM). RdDM requires the specialized RNA polymerase V (Pol V), and the key unanswered question is how Pol V is first recruited to new target sites without pre-existing DNA methylation. We find that Pol V follows and is dependent on the recruitment of an AGO4-clade ARGONAUTE protein, and any siRNA can guide the ARGONAUTE protein to the new target locus independent of pre-existing DNA methylation. These findings reject long-standing models of RdDM initiation and instead demonstrate that siRNA-guided ARGONAUTE targeting is necessary, sufficient and first to target Pol V recruitment and trigger the cycle of RdDM at a transcribed target locus, thereby establishing epigenetic silencing.
Abstract
This study finds that siRNA-guided ARGONAUTE first recruits polymerase V to new target sites without pre-existing DNA methylation and triggers the cycle of RdDM at the target sites, thereby establishing epigenetic silencing.
Related collections
Most cited references57

- Record: found
- Abstract: found
- Article: found
BEDTools: a flexible suite of utilities for comparing genomic features
- Record: found
- Abstract: not found
- Article: not found
Dynamics and function of DNA methylation in plants
- Record: found
- Abstract: found
- Article: not found