- Record: found
- Abstract: found
- Article: not found
Enzyme-Instructed Intracellular Peptide Assemblies.
Abstract
Higher-order or supramolecular protein assemblies, usually regulated by enzymatic reactions, are ubiquitous and essential for cellular functions. This evolutionary fact has provided a rigorous scientific foundation, as well as an inspiring blueprint, for exploring supramolecular assemblies of man-made molecules that are responsive to biological cues as a novel class of therapeutics for biomedicine. Among the emerging man-made supramolecular structures, peptide assemblies, formed by enzyme reactions or other stimuli, have received most of the research attention and advanced most rapidly.In this Account, we will review works that apply enzyme-instructed self-assembly (EISA) to generate intracellular peptide assemblies for developing a new kind of biomedicine, especially in the field of novel cancer nanomedicines and modulating cell morphogenesis. As a versatile and cell-compatible approach, EISA can generate nondiffusive peptide assemblies locally; thus, it provides a unique approach to target subcellular organelles with exceptional cell selectivity. We have arranged this Account in the following way: after introducing the concept, simplicity, and uniqueness of EISA, we discuss the EISA-formed intracellular peptide assemblies, including artificial filaments, in the cell cytosol. Then, we describe the representative examples targeting subcellular organelles, such as mitochondria, endoplasmic reticulum, Golgi apparatus, lysosomes, and the nucleus, by enzyme-instructed intracellular peptide assemblies for potential cancer therapeutics. After that, we highlight the recent exploration of the transcytosis of peptide assemblies for controlling cell morphogenesis. Finally, we provide a brief outlook of enzyme-instructed intracellular peptide assemblies. This Account aims to illustrate the promise of EISA-generated intracellular peptide assemblies in understanding diseases, controlling cell behaviors, and developing new therapeutics from a class of less explored molecular entities, which are substrates of enzymes and become building blocks of self-assembly after the enzymatic reactions.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: found
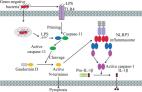
Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors

- Record: found
- Abstract: found
- Article: found