- Record: found
- Abstract: found
- Article: found
Transcriptional repression of frequency by the IEC-1-INO80 complex is required for normal Neurospora circadian clock function
Read this article at
Abstract
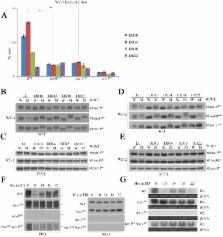
Rhythmic activation and repression of the frequency ( frq) gene are essential for normal function of the Neurospora circadian clock. WHITE COLLAR (WC) complex, the positive element of the Neurospora circadian system, is responsible for stimulation of frq transcription. We report that a C2H2 finger domain-containing protein IEC-1 and its associated chromatin remodeling complex INO80 play important roles in normal Neurospora circadian clock function. In iec-1 KO strains, circadian rhythms are abolished, and the frq transcript levels are increased compared to that of the wild-type strain. Similar results are observed in mutant strains of the INO80 subunits. Furthermore, ChIP data show that recruitment of the INO80 complex to the frq promoter is IEC-1-dependent. WC-mediated transcription of frq contributes to the rhythmic binding of the INO80 complex at the frq promoter. As demonstrated by ChIP analysis, the INO80 complex is required for the re-establishment of the dense chromatin environment at the frq promoter. In addition, WC-independent frq transcription is present in ino80 mutants. Altogether, our data indicate that the INO80 complex suppresses frq transcription by re-assembling the suppressive mechanisms at the frq promoter after transcription of frq.
Author summary
Circadian clocks organize inner physiology to anticipate changes in the external environment. These clocks are controlled by the oscillation of central clock proteins which form the central oscillator. Transcriptional regulation is a critical step in the regulation of the oscillation of these core proteins. In eukaryotes, chromatin remodeling is a common mechanism to regulate gene transcription by conquering or establishing nucleosomal barriers for the transcription machinery. Here, we showed that a C2H2 finger domain-containing protein IEC-1 and its associated chromatin remodeling complex INO80 are required for transcriptional repression of the core clock gene frq in the Neurospora circadian system. Moreover, the activator WHITE COLLAR (WC) complex is responsible for the transcriptional activation of frq; thus, considering the different timing of the transcriptional activation and suppression of frq, there must be a mechanism that coordinates the two opposite processes. We also demonstrated that the WC-mediated open state of the frq promoter facilitates the binding of INO80 to this region, which prepares for subsequent transcriptional suppression. Collectively, our data provide novel insights into the regulation of the frq gene and the circadian clock.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Nascent transcript sequencing visualizes transcription at nucleotide resolution.
- Record: found
- Abstract: found
- Article: not found
Time zones: a comparative genetics of circadian clocks.
- Record: found
- Abstract: found
- Article: not found