- Record: found
- Abstract: found
- Article: found
NC2 complex is a key factor for the activation of catalase-3 transcription by regulating H2A.Z deposition
Read this article at
Abstract
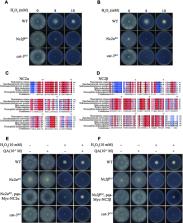
Negative cofactor 2 (NC2), including two subunits NC2α and NC2β, is a conserved positive/negative regulator of class II gene transcription in eukaryotes. It is known that NC2 functions by regulating the assembly of the transcription preinitiation complex. However, the exact role of NC2 in transcriptional regulation is still unclear. Here, we reveal that, in Neurospora crassa, NC2 activates catalase-3 ( cat-3) gene transcription in the form of heterodimer mediated by histone fold (HF) domains of two subunits. Deletion of HF domain in either of two subunits disrupts the NC2α–NC2β interaction and the binding of intact NC2 heterodimer to cat-3 locus. Loss of NC2 dramatically increases histone variant H2A.Z deposition at cat-3 locus. Further studies show that NC2 recruits chromatin remodeling complex INO80C to remove H2A.Z from the nucleosomes around cat-3 locus, resulting in transcriptional activation of cat-3. Besides HF domains of two subunits, interestingly, C-terminal repression domain of NC2β is required not only for NC2 binding to cat-3 locus, but also for the recruitment of INO80C to cat-3 locus and removal of H2A.Z from the nucleosomes. Collectively, our findings reveal a novel mechanism of NC2 in transcription activation through recruiting INO80C to remove H2A.Z from special H2A.Z-containing nucleosomes.
Related collections
Most cited references63

- Record: found
- Abstract: found
- Article: found
The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles
- Record: found
- Abstract: found
- Article: not found
The general transcription machinery and general cofactors.

- Record: found
- Abstract: found
- Article: found